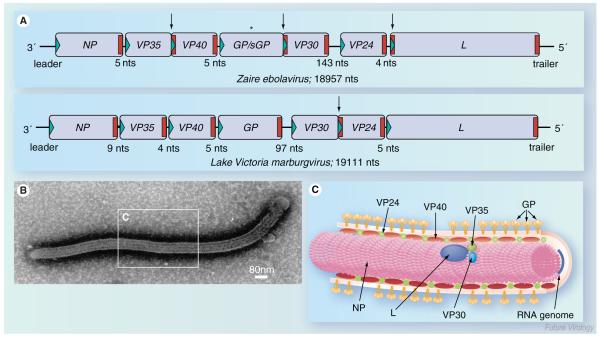
Figure 1. Filovirus genome organization and structure.
(A) Schematic diagram of Ebola virus (EBOV) and Marburg virus (MARV) genomes. The genes are depicted as boxes and nontranscribed regions as black bars (leader, trailer and intergenic regions). Transcription start signals are depicted as green triangles and stop signals as red bars. Gene overlaps are marked by arrows. The mRNA editing site within the EBOV GP gene is indicated by an asterisk. The length of the intergenic regions is shown below the scheme. Note that the EBOV VP24 gene contains two transcription stop signals. (B) Electron micrograph of an EBOV particle. (C) Schematic presentation of EBOV structure. The RNA genome is encapsidated by nucleocapsid proteins NP, VP35, VP30 and L. VP40 and VP24 are matrix proteins. GP trimers are inserted into the viral membrane.
GP: Glycoprotein; L: RNA-dependent RNA polymerase; NP: Nucleoprotein; VP: Viral protein.