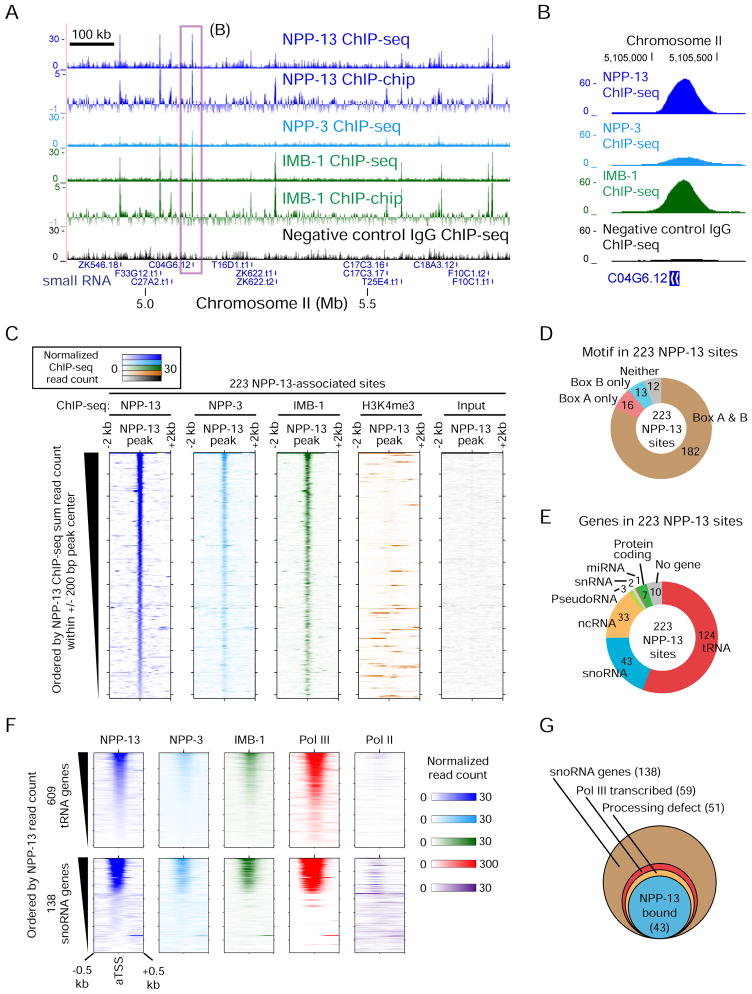
Figure 2. Nuclear pore proteins associate with small RNA genes in C. elegans.
(A) ChIP-seq and ChIP-chip profiles of nuclear pore proteins. For ChIP-seq, read counts normalized by average base coverage are shown (also in B, C and F). For ChIP-chip, MA2C scores are shown. The box indicates a region shown in detail in (B). Only small RNA genes are shown under tracks. Signals over the y-axis range are omitted. The specificity of the NPP-13 antibody is validated in Figure S2A.
(B) Nucleoporin ChIP-seq signals at snoRNA gene C04G6.12.
(C) Colorimetric representation of per-base ChIP-seq read count at 223 NPP-13-associated sites. Orientation is according to the genome coordinate.
(D) The number of NPP-13-associated sites harboring Box A or B motifs in the 200 bp region centered on the peak summit.
(E) The class of genes found in the 1 kb region centered on the summit of NPP-13-associated sites. The search was performed first for non-protein coding RNA genes, then for protein-coding genes.
(F) Colorimetric representation of nucleoporin and RNA polymerase ChIP-seq read counts for all annotated tRNA and snoRNA genes.
(G) Summary of 138 snoRNA genes. Of 51 genes showing RNA processing defects upon NPP-13 depletion, 43 are bound by NPP-13 at the level exceeding our peak calling threshold. The remaining 8 genes are still associated with NPP-13 but below the cutoff.