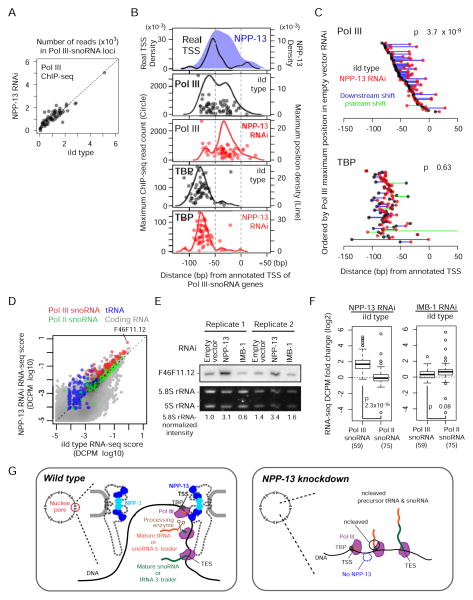
Figure 4. NPP-13 knockdown alters Pol III distribution on snoRNA genes and increases tRNA and snoRNA levels.
(A) Pol III ChIP-seq read counts in NPP-13 knockdown versus wild-type embryos at 59 Pol III-snoRNA genes. Reads overlapping with regions corresponding to -100 bp to the transcription termination site are normalized by average base coverage and plotted.
(B) Positions of Pol III and TBP ChIP-seq signal maxima in wild-type (empty RNAi vector) or NPP-13 knockdown embryos are shown as in Figure 3E. For comparison, the positions of real TSSs and wild-type NPP-13 ChIP-seq maxima used in Figure 3E are shown.
(C) Trajectories of Pol III and TBP maxima shift upon NPP-13 knockdown. Circles represent wild-type and NPP-13-knockdown Pol III maximum positions. Lines connect wild type and NPP-13 knockdown circles of the same gene. P-value is calculated by paired t-test for increased downstream shift.
(D) RNA abundance of NPP-13 knockdown versus wild-type embryos determined by RNA-seq and shown as a DCPM (Depth of Coverage Per base per Million reads) score. Each circle represents a single transcript. Transcripts with DCPM value smaller than 10−5 are not shown.
(E) Northern blotting of snoRNA F46F11.12 (also indicated in D) confirms the increased RNA abundance after NPP-13 knockdown.
(F) Boxplots indicate the log2 fold change of the RNA abundance upon NPP-13 knockdown (left) or IMB-1 knockdown (right). Student’s t-test is used for the statistical analysis. Parentheses indicate the number of transcripts annotated in each category.
(G) A model depicting that the nuclear pore serves as a hub for Pol III transcription and subsequent processing. In wild type C. elegans (left), scaffold nucleoporins associate with snoRNA and tRNA genes that undergo high levels of Pol III transcription. The nascent transcripts are processed by a yet unidentified endonuclease, perhaps in a co-transcriptional manner. Given the stability of NPP-13 and NPP-3 at the nuclear pore, this series of events likely occur in the pore channel although this is still our speculation. In the absence of NPP-13 (right), Pol III can still transcribe the target genes, but the transcripts fail to undergo processing, resulting in producing uncleaved precursor snoRNAs and tRNAs. See also Figure S3 for a size-scaled model.