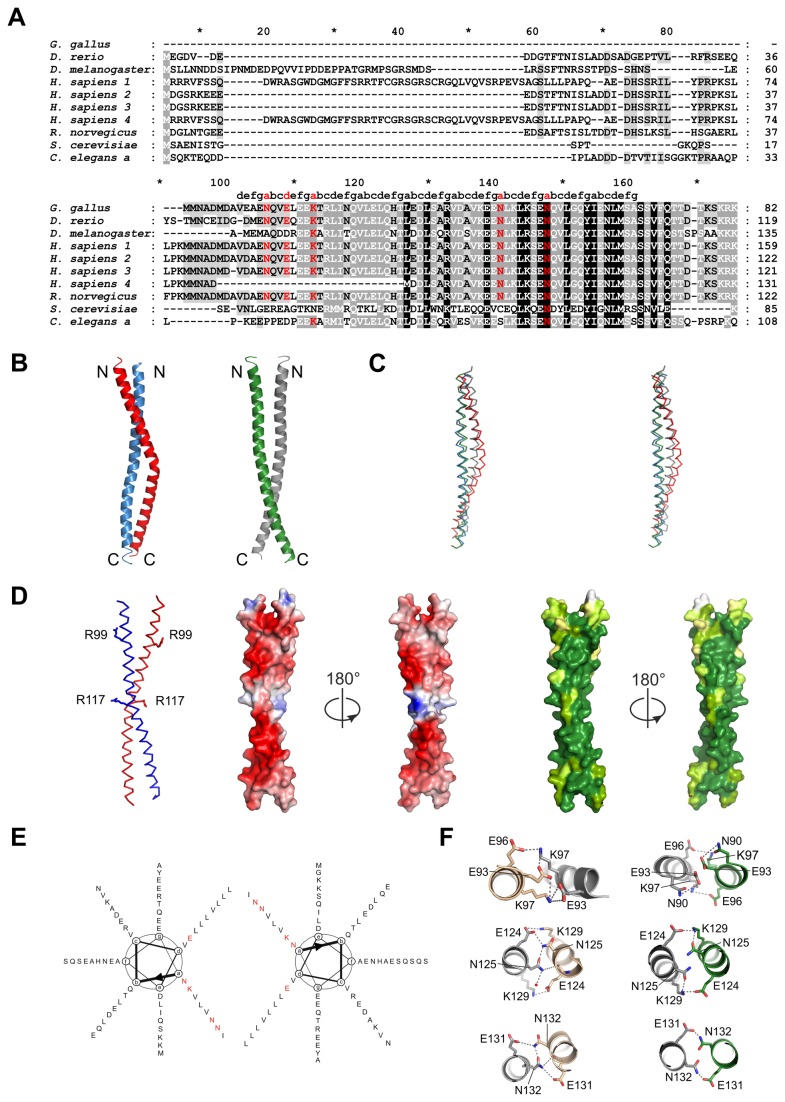
Figure 1. Structure of the SCOC coiled coil domain.
(A) Sequence alignment of human SCOC isoforms and homologues was done with T-Coffee [35]. Accession numbers are: G. gallus: NP_001108324.1, D. rerio: NP_001071027.1, D. melanogaster: NP_651552.1, H. sapiens 1: NP_001146956.1, H. sapiens 2: Q9UIL1-2, H. sapiens 3: Q9UIL1-3, H. sapiens 4: Q9UIL1-4, R. norvegicus: NP_001013253.1, S. cerevisiae: NP_796378.3, C. elegans a: NP_001022752.1. Coiled coil heptad positions were assigned with TWISTER [29]. Non-canonical polar amino acids at core positions a and d are colored red. (B) Cartoon representation of the two SCOC dimers AB and CC’. Molecule A is colored red, B blue, C green and C’ grey. (C) Stereo view of dimers overlay. Same coloring as in (B). (D) Left panel shows a ribbon representation of dimer AB. Residues R99 and R117 are drawn as stick models. Electrostatic surface potential is shown for dimer AB in two orientations. Blue shows positive charge and red corresponds to negative charge. The first figure of the electrostatic surface potential is shown in the same orientation as the ribbon representation. The two right panels show the surface conservation of SCOC. Dark green correspond to strong conservation and lighter green shades represent less conserved regions. The first conservation figure is shown in the same orientation as the ribbon representation. Sequence alignments were done with T-Coffee and analysis of the degree of conservation was done with AMAS [36]. (E) Helical wheel diagram of the SCOC dimer. Polar and charged core residues are marked red. (F) Detailed views on molecular interactions of the non-canonical polar core positions of dimer AB (left panel) and dimer CC’ (right panel). Core residue E93, K97, N125 and N132, which were used for mutagenesis studies are stabilized through a network of hydrogen bonds and salt bridges as shown. Figures were prepared with PyMol [22].