Figure 1.
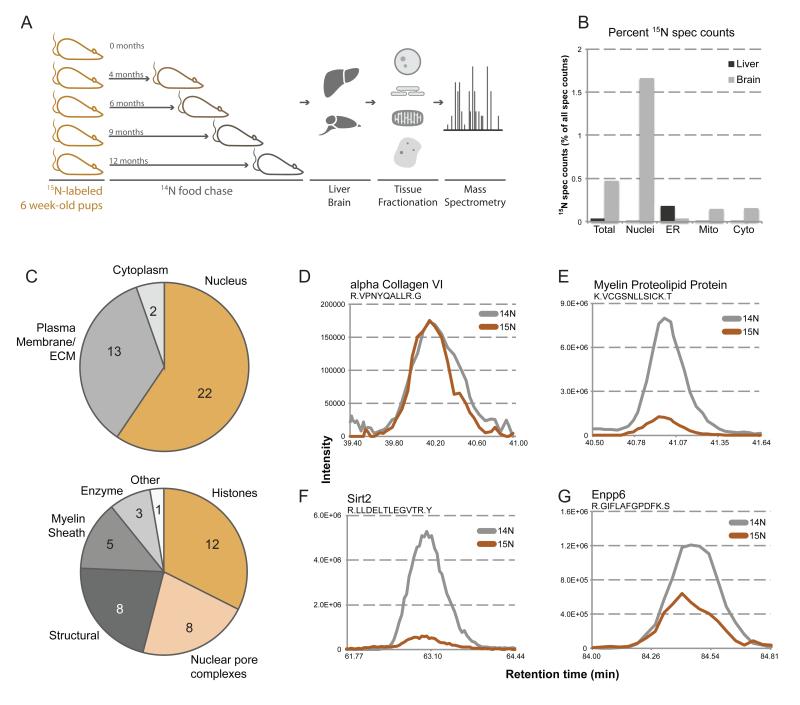
Discovery of new members of the long-lived proteome. (A) Pulse-chase labeling of whole rats. Depicted is a schematic of the pulse-chase labeling procedure. Litters of rats were fully 15N-labeled through feeding a 15N diet starting from a previous generation. Fully labeled rats were then switched to a normal 14N diet (chase) at 6 weeks post-natal, and sacrificed a 0, 4, 6, 9, and 12 months post-chase. Tissues were harvested, fractionated, and analyzed by MS (B) Tissue localization of long-lived proteins. 15N spectral counts were calculated and plotted as a percentage of total spectral counts in each fraction of liver (grey) and brain (black) tissues from an animal 6-months post-chase. (C) Cellular localization and processes of long-lived proteins. Identified long-lived proteins were sorted by subcellular localization (upper) and cellular process (lower), and plotted as a pie chart, inset numbers representing the number of proteins per localization. (D-G) MS1 traces of representative peptides. Aligned elution profile MS1 traces are plotted for representative peptides for alpha collagen VI (D), myelin proteolipid protein (E), Sirt2 (F), and Enpp6 (G), with 15N signal in orange and 14N signal in grey. See also Figure S1-3.