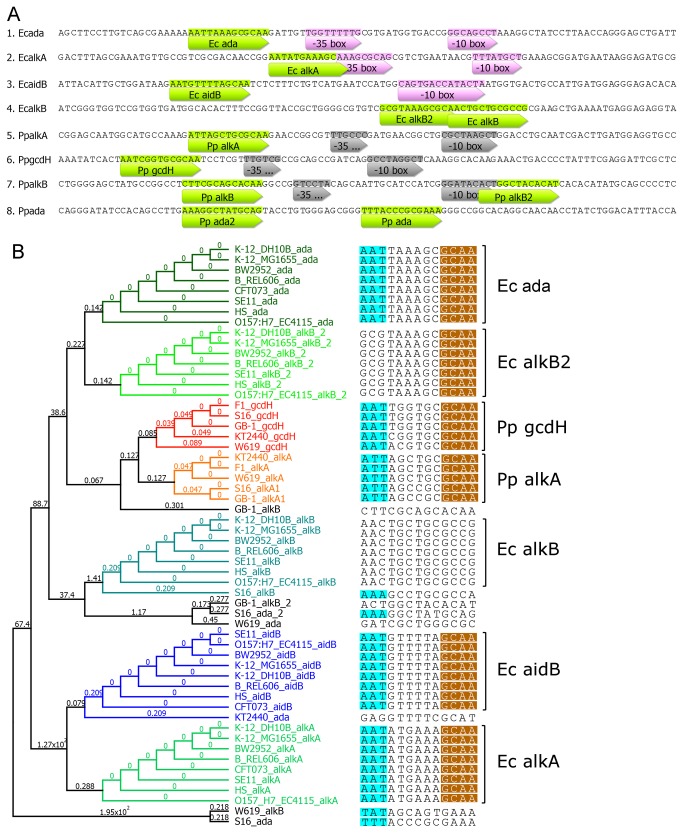
Figure 3. The consensus sequences responsible for Ada protein specific interaction (A) located in promoter sequences (up to -100 nucleotide residues relative to ATG start codons) of E. coli ada, alkA, and aidB.
Additionally, the predicted consensuses in promoter regions of E. coli alkB and P. putida alkA, gcdH, alkB, and ada genes are shown (green bars). Also, the putative -35 and -10 boxes responsible for RNA polymerase interaction are marked in pink (retrieved from RegulonDB, http://regulondb.ccg.unam.mx) or grey (SoftBerry BPROM prediction tool, http://linux1.softberry.com). The phylogenetic tree (B) shows relative conservation between depicted consensus sequences based on UPGMA Geneious Tree Builder, A box – light blue, B box – brown, numbers indicate substitutions per site (for details see Materials and Methods).