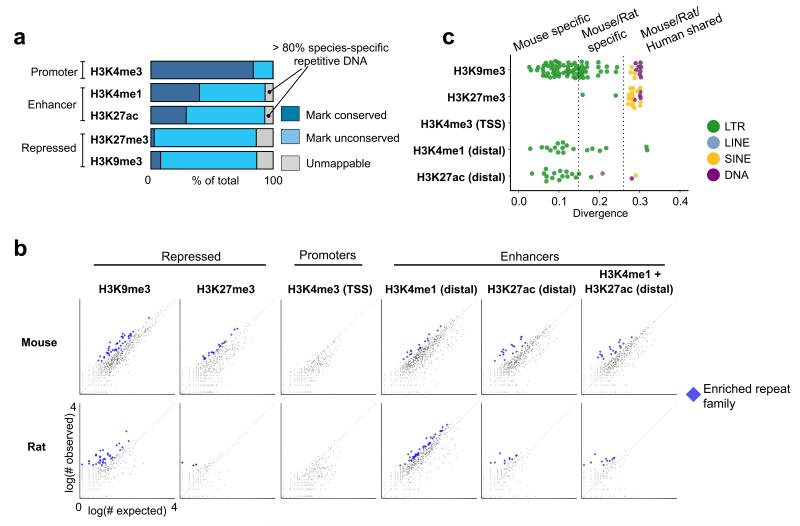
Figure 2. Comparison between mouse and rat reveals over abundance of species specific ERVs in enhancer regions.
(a) Rat TSC ChIP-Seq defined regulatory elements were mapped to their orthologous position in the mouse genome. If the same histone mark was present in mouse, then the element was considered epigenetically conserved. For unconserved elements, we further distinguished whether the genomic DNA was mappable to the other genome, or derived from species-specific sequence. Each category is represented as a fraction of the total number of elements in the ChIP-Seq dataset (dark blue: rat-mouse conserved elements, light blue: unconserved regions gray: unmappable elements). In both species, the unmappable regions were predominantly composed of species-specific TEs. (b) We deduced whether the frequency of any type of TE was enriched within each class of regulatory element. Each point represents a single TE family, composed of up to several thousand copies genome-wide. For each family, the number of individual copies observed residing within a set of regulatory elements (Y axis) is plot against a random expectation (X axis). Significantly overrepresented families are indicated in blue. (c) Overrepresented mouse TE families from (b) are plot against the average nucleotide divergence of their individual copies versus the consensus sequence, which is a proxy for the evolutionary age of the TE. Each point is colored based on the class of TE. Divergence measurements representing the distance between mouse/rat and mouse/human are depicted by dotted lines. ERVs: endogenous retroviruses; TE: transposable elements.