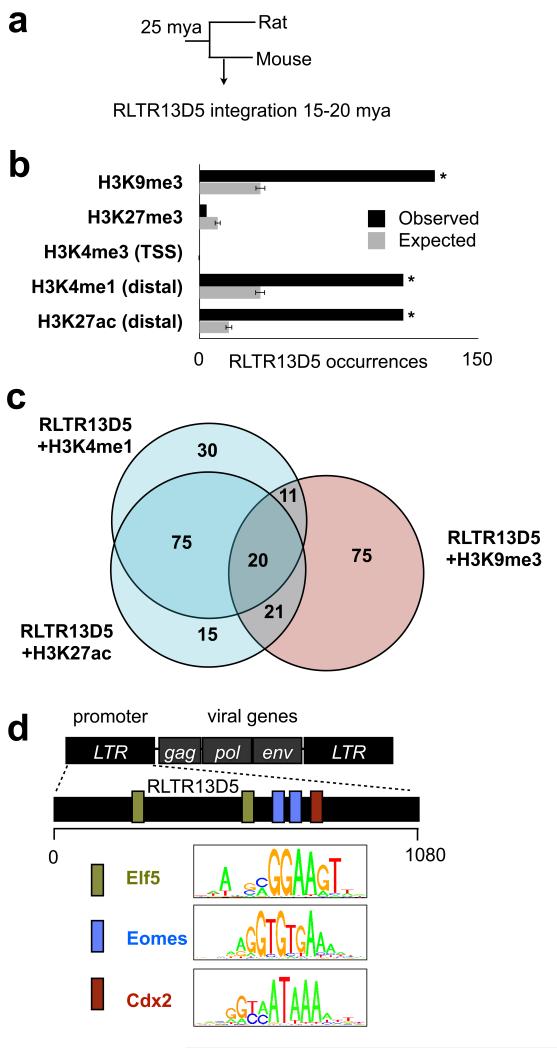
Figure 3. Mouse-specific ERV RLTR13D5 is highly enriched within placental enhancers.
(a) Phylogenetic tree indicating approximate RLTR13D5 integration time in mouse genome. (b) Examination of all 608 instances of RLTR13D5 shows that this family is highly enriched within the enhancer marks H3K4me1 (Bonferroni P = 4.5 × 10−29, binomial test) and H3K27ac (P = 4.2 × 10−57) as well as for the repressive mark H3K9me3 (P = 1.5 × 10−36). This is illustrated in a barplot comparing the observed number of RLTR13D5 copies within a histone modification to the random expectation. The random expectation is displayed as the average over 1000 randomized datasets, and error bars indicate standard deviation. (c) Venn Diagram showing that RLTR13D5 instances containing the H3K4me1 and H3K27ac enhancer marks are distinct from those containing the repressive mark H3K9me3. (d) Diagram of RLTR13D5, whose sequence originally derives from a long terminal repeat (LTR) segment of an ERV. The RLTR13D5 consensus sequence harboring predicted binding sites for Eomes, Cdx2, and Elf5, which are depicted by colored boxes across the 1080 bp-long consensus sequence. Uniprobe motifs used to scan the sequence are shown in the legend.