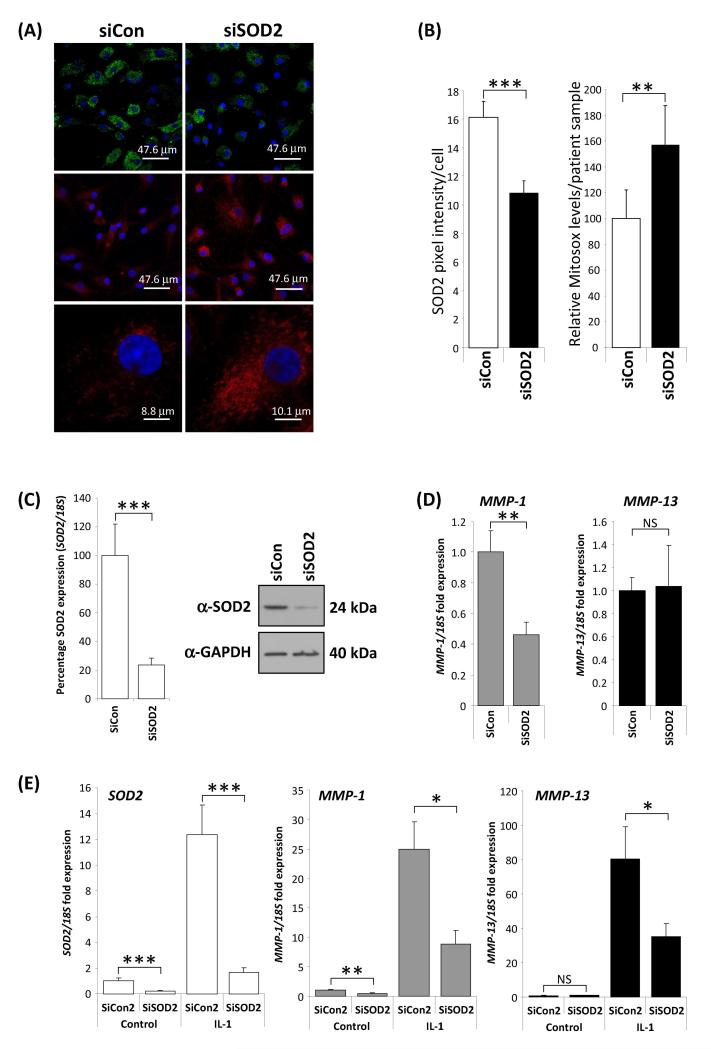
Figure 5.
Functional consequences of SOD2 depletion. A and B, SOD2 depletion (green) by RNAi led to a significant increase in MitoSOX™ Red mitochondrial staining as determined by confocal microscropy. Data are taken from four patient chondrocyte preparations. Images were quantified using ImageJ software on a per cell basis from random field images. P values were calculated using a Student’s t-test, where ** represents P<0.01 and *** P<0.001 C-E, Depletion of SOD2 leads to reduction in collagenase gene expression. C, transfection of primary human articular chondrocytes (HAC) from OA patients with siRNA to SOD2 (siSOD2) for 48 hours under basal conditions (24 hour serum free) compared to transfection with an equivalent amount (100nM) of a non-targeting control siRNA (siCon) resulted in a highly significant approximately 80% reduction in SOD2 RNA level as determined by realtime RT-PCR. Concomitantly a substantial reduction in SOD2 protein expression was measured by immunoblotting (an anti-GAPDH antibody was used as a protein loading control). D, MMP-1 and MMP-13 basal expression levels were measured as described above. SOD2 RNAi resulted in a significant reduction in MMP-1 but no change in the levels of MMP-13. E, interleukin-1 (IL-1, 0.05 ng/ml) stimulation for 24 hours induced the expression of SOD2, MMP-1 and MMP-13, however the induced-expression of all three genes was significantly reduced when SOD2 was depleted by RNAi (siSOD2). Throughout RNA expression levels were normalised to the 18S gene and data plotted as fold over control levels. Data presented are from a single experiment (n=8) using cells from one donor. The experiment was performed three times using different patient cell populations. P values were calculated using the Mann-Whitney U-test, where * represents P<0.05, ** P<0.01 and *** P<0.001. Error bars shown are SEM.