Figure 1.
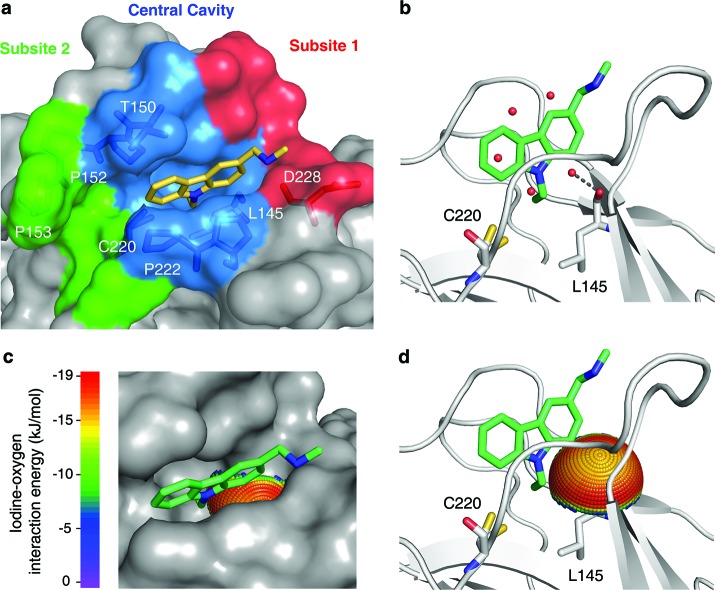
Mutation-induced cavity in p53 Y220C. (a) Molecular surface of the p53 mutant Y220C bound to the carbazole derivative PhiKan083 (PDB code 2VUK).19 The ligand is shown as a yellow stick model. The mutation-induced cavity can be subdivided into three parts: (i) a deep but narrow central cavity, colored in blue, which is occupied by the carbazole ring; (ii) an open rather shallow subsite 1, colored in red; and (iii) subsite 2, colored in green, which is flanked by several prolines and main-chain oxygens. The latter is not occupied by PhiKan083. (b) Ribbon diagram showing details of the binding mode of PhiKan083, in particular the role of Leu145 at the bottom of the central cavity.19 Also shown are structural water molecules in the ligand-free structure (PDB code 2J1X)18 that are displaced upon ligand binding. One of these water molecules sits at the bottom of the predominantly hydrophobic central cavity and forms a hydrogen bond with the main-chain oxygen of Leu145. (c and d) Potential for halogen-bond interactions with the carbonyl oxygen of Leu145. The iodine-interaction energy sphere is plotted onto the carbonyl oxygen of Leu145 in the Y220C-PhiKan083 structure, showing that this oxygen at the bottom of the central cavity is poised for interaction with iodine-containing ligands with maximum binding energy. Blue regions have the lowest energy; red regions have the highest energy (see Supporting Information for details).