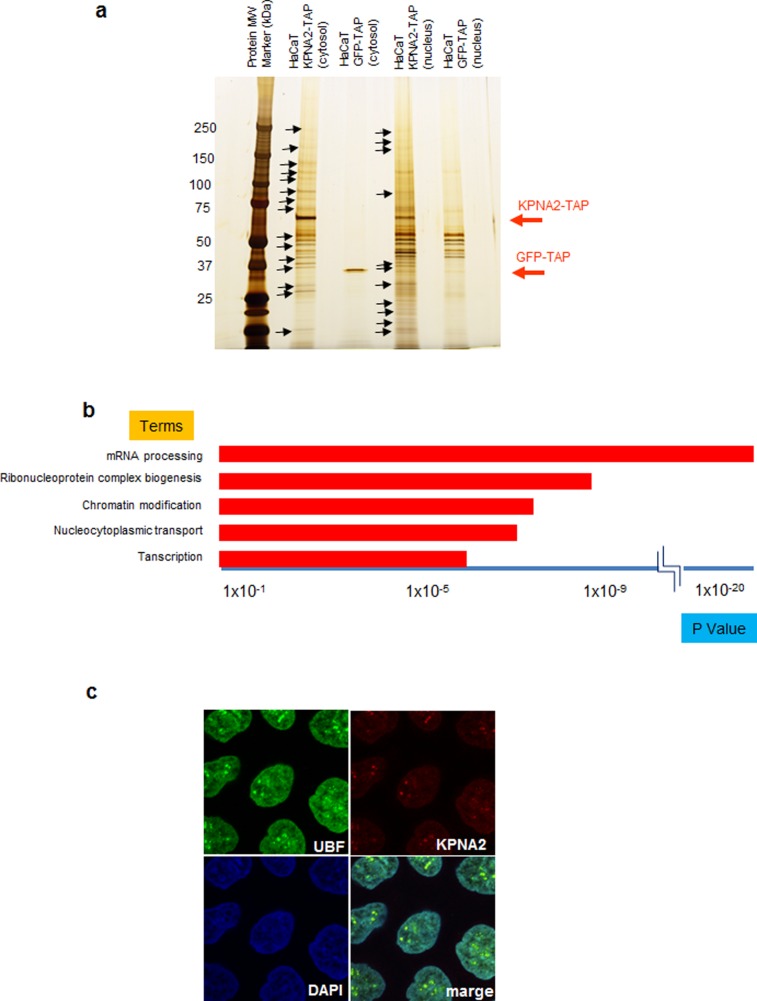
Figure 3. Detection and analysis of proteins that interact with KPNA2 and localization of KPNA2 in the nucleolus.
Proteins that interact with KPNA2 in the cytoplasm and nucleus were purified using the TAP method and detected by silver staining. Proteins marked with arrows were analyzed by LC/MS/MS. HaCaT cells expressing GFP-TAP were used to detect nonspecific interactions. a) The results of LC/MS/MS were analyzed by pathway analysis using reactome (http://www.reactome.org). The categories of “mRNA processing”, “ribonucleoprotein complex biogenesis”, “chromatin modification,” and “transcription” were the most significantly represented pathways. b) Immunohistochemistry revealed KPNA2 co-localization with UBF, a nucleolar marker.