Figure 5. Validation of top candidate insulators in conventional drug-resistant colony assay.
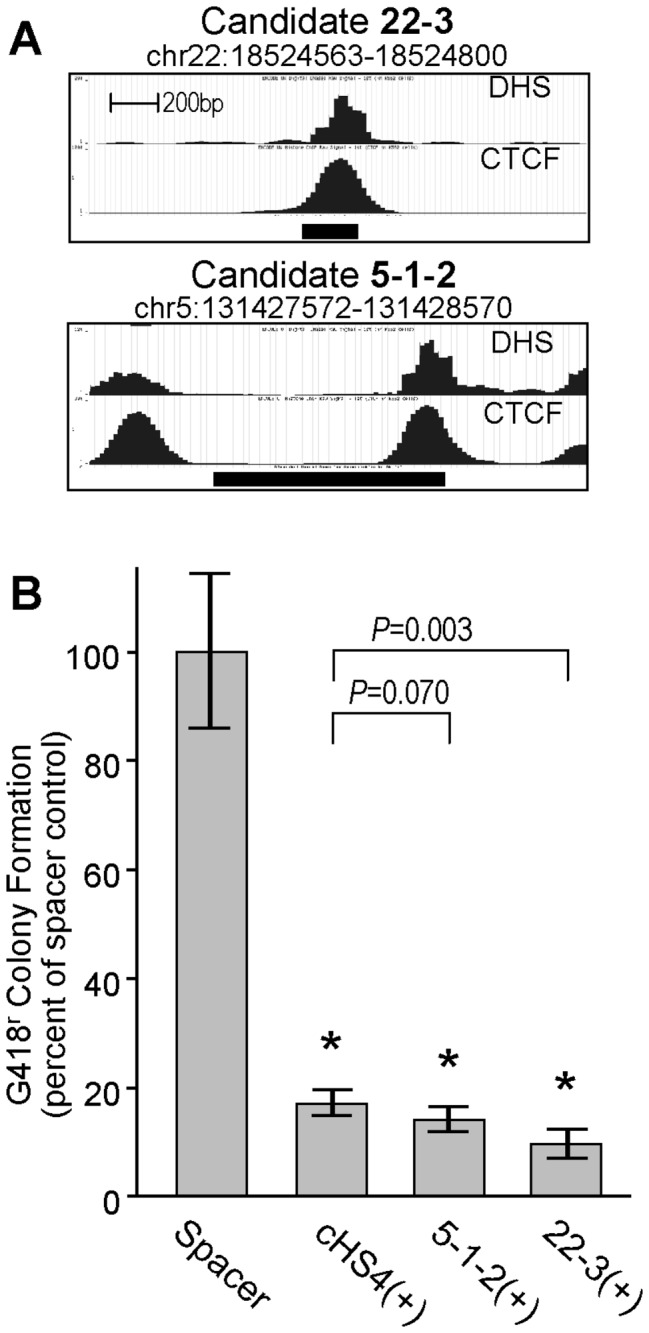
(A) Chromatin profiling for top two insulator candidates. The raw sequence tags are shown for the DHS and CTCF-binding activities over the 2 kb regions centered around the top insulator candidates 22-3 and 5-1-2 (dark horizontal bars indicate locations of candidate sequences). (B) Insulator activity. The top two candidate insulator elements 5-1-2 and 22-3, the 1.2 kb cHS4 insulator, or a neutral spacer (308 bp fragment from the bacterial drug resistance gene Zeo), were inserted into the reporter plasmid pJC5-4/P4-P2K (Figure 1A) in the (+) orientation and analyzed for colony formation under G418 selection (1000 µg/mL). Histograms represent the average ± standard deviation from a total of 4 samples in 2 independent experiments, and are reported as a percentage of the average colony formation obtained with the Zeo control (set at 100%). P values presented for comparisons between candidate elements versus the cHS4 positive control are based on t-test. * indicate P<0.001 for the cHS4 and all candidate elements versus the spacer control (t-test).
