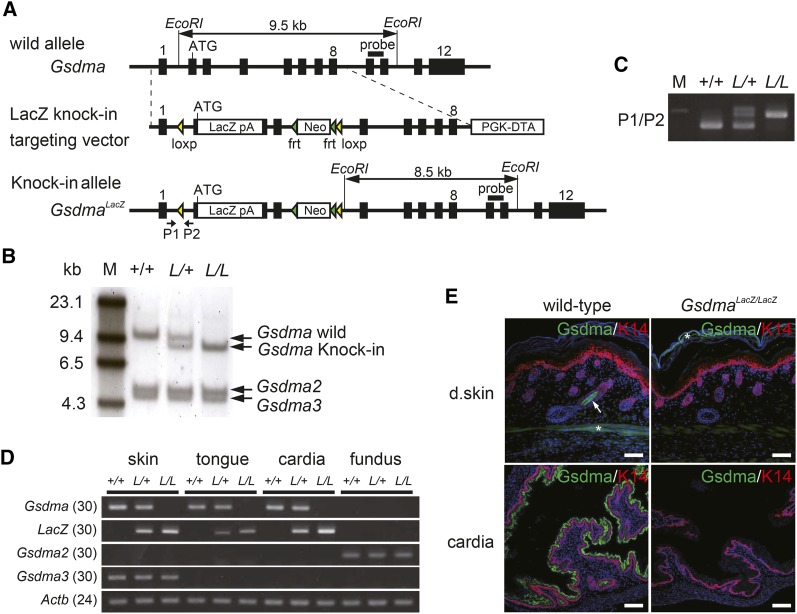
Figure 2.
Generation and validation of the Gsdma KO mouse. (A) Schematic diagrams of wild-allele, LacZ knock-in targeting vector, and knock-in allele of the Gsdma gene. Exons are indicated as black boxes. loxP and frt sites are indicated as yellow and green triangles, respectively. The LacZ-pA fragment was inserted into the start codon in exon 2. EcoRI fragments and the probe for Southern blot analysis are indicated by horizontal bars with double arrows and horizontal bars. Location of primers used for genotyping are indicated by allows and labeled P1 and P2. (B) Southern blot analysis of EcoRI digested genomic DNA from wild-type (+/+), heterozygous (L/+), and homozygous (L/L) mice for the LacZ knock-in allele. The probe located in exon 9−10 detects 9.5- and 8.5-kb fragments of the wild and knock-in allele, respectively. The DNA fragments containing Gsdma2 and Gsdma3 are intact. M indicates a marker. (C) PCR analysis of genomic DNA from mice with each genotype. Genotype was determined by PCR using primers P1 and P2. This yielded fragments of 410 and 530 bp for the wild and knock-in allele, respectively. (D) Semiquantitative reverse-transcription PCR analysis of Gsdma, Gsdma2, Gsdma3, and LacZ genes using cDNAs prepared from skin, tongue, cardia, and gastric fundus of each genotype. Actin-beta (Actb) was amplified as a control. The number of PCR cycles is shown in parentheses. (E) Immunohistological detection of Gsdma protein in skin and stomach of wild-type and GsdmaLacZ/LacZ mice at E18.5. K14 was used as a marker for the basal cell layer. An arrow and asterisks indicate specific and nonspecific signal, respectively. Scale bar indicates 50 μm.