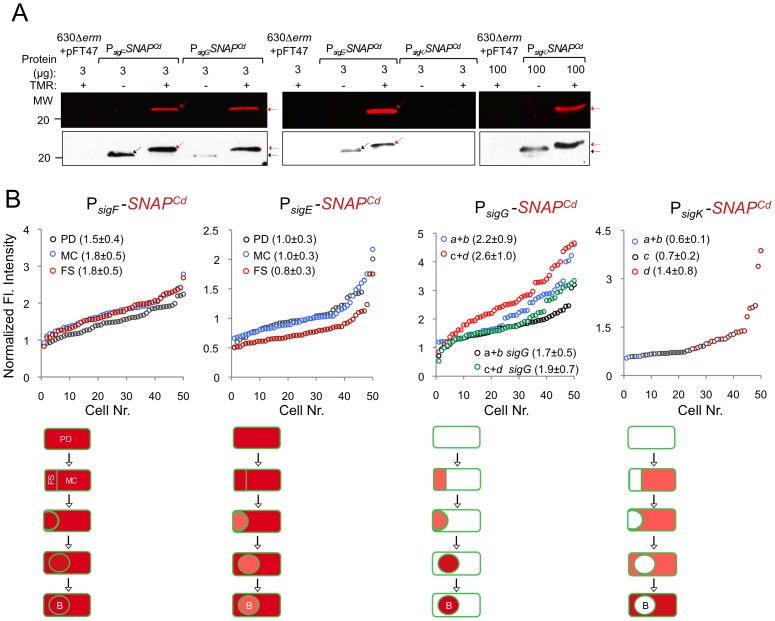
Figure 5. Quantitative analysis of sigF, sigE, sigG and sigK expression during sporulation.
(A) Whole cell extracts were prepared from derivatives of strain 630Δerm bearing the indicated plasmids or fusions, immediately after labeling with TMR-Star, indicated by the “+” sign (the “−” sign indicates control, unlabeled samples). The indicated amount of total protein was resolved by SDS-PAGE, and the gels scanned on a fluorimager (top) or subject to immunoblotting with anti-SNAP antibodies (bottom). Black and red arrows point to unlabeled or TMR-Star-labeled, respectively, SNAP. Strain 630Δerm carrying pFT47 (empty vector) was used as a negative control for SNAP production. The position of molecular weight markers (in kDa) is indicated. (B) Quantitative analysis of the fluorescence (Fl.) intensity in different cell types of the reporter strains for sigF, sigE, sigG and sigK transcription, as indicated. The numbers in the legend represent the average ± SD of fluorescence intensity for the cell class considered (n = 50 cells analysed for each morphological cell class). The data shown are from one experiment, representative of three independent experiments. Schematic representation of the deduced spatial and temporal pattern of transcription (with darker red denoting increased transcription) is shown for each transcriptional fusion. The cell membrane is represented in green. PD, pre-divisional cell; MC, mother cell; FS, forespore; B, phase bright spore; a to d: sporulation classes as defined in Figure 1.