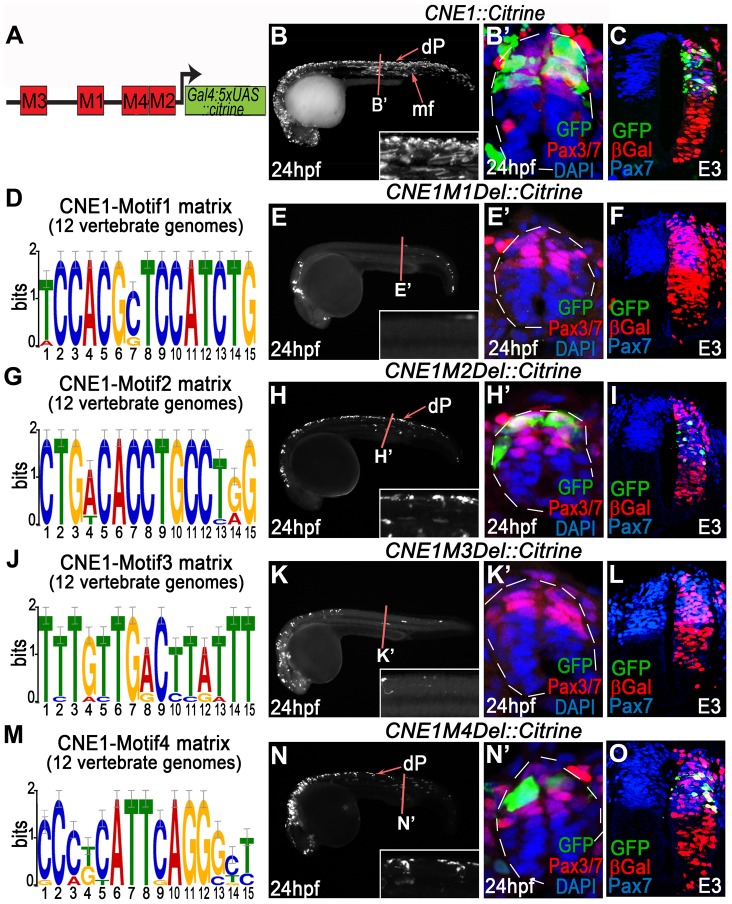
Figure 3. Functional dissection of CNE1 enhancer activity.
(A) Schematic outlining the organisation of motifs within CNE1. (B, B′) CNE1 transient transgenic zebrafish embryos recapitulate pax3a expression across the AP axis of the CNS, including the spinal cord (n = 66/67). (C) Zebrafish CNE1 sequence specifically drives transgene expression in the Pax3/7 domain of the chick spinal cord, despite widespread transfection of LacZ across the DV axis of the tissue (n = 10/12). (D) Matrix representing the degree of Motif1 conservation across 12 vertebrate genomes. (E, E′) Deletion of Motif1 results in a complete loss of CNE1 activity in zebrafish spinal cord at trunk level (n = 0/22, p<0.0001), however the enhancer remains active in the anterior CNS (n = 22/22) and the most posterior region of the spinal cord (n = 8/22). (F) Loss of Motif1 greatly reduces CNE1 activity in the chick neural tube (n = 1/8, p = 0.0019). Motif2 (G), is not required for CNE1 activity in the zebrafish (H, H′) (n = 37/38) or chick (I) (n = 4/6) spinal cord. Loss of Motif 3 (J) reduces CNE1 mediated transcription across the AP axis of the zebrafish CNS (K, K′) (n = 0/26, p<0.0001) and precludes activity in the chick spinal cord (L) (n = 0/5, p = 0.003). Deletion of Motif4 (M) does not significantly alter the activity of CNE1 in zebrafish (N, N′) (n = 20) or chick (O) (n = 4/5).