FIGURE 1.
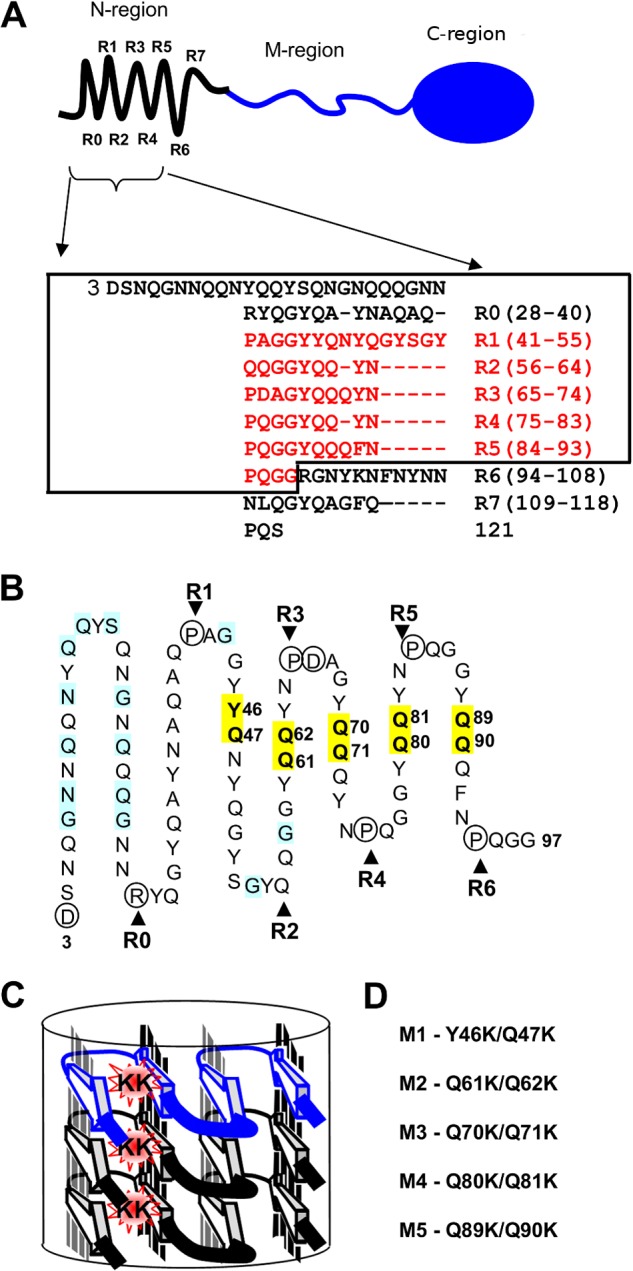
Localization of mutated Sup35 residues regarding the structural model of the N-terminal domain. A, top, schematic representation of Sup35p domain organization: the N-terminal region (1–123 amino acids) is shown with an array of eight repeats (R0–R7), discovered with the T-REKS program, the M-region (124–245 amino acids), C-terminal region (246–685 amino acids) has a globular-fold of the GTP-binding domain essential for robust translation termination. Bottom, new assignment of the Sup35p tandem repeats (T-REKS), which includes a previous one (8) colored in red. B, sequence of the N-terminal domain (the rectangle within the panel A) arranged in a serpentine with linear regions and bends, corresponding to putative β-strands and loop regions, respectively. The circled residues tend to localize to the loop regions (proline and charged residues). Positions where substitutions with charged residues lead to the PNM phenotype in previous reports (19, 36, 48) are highlighted in blue. Residues highlighted in yellow are mutated to lysines in this study. C, a model for the super-pleated β-structure that demonstrates how the introduction of charged residues in the β-strand region destabilizes the fibril by the electrostatic repulsion of same-sign charges along the fibril axis. D, sup35 mutations studied in this work (numbering of the mutations corresponds to the repeat number shown above).
