FIGURE 1.
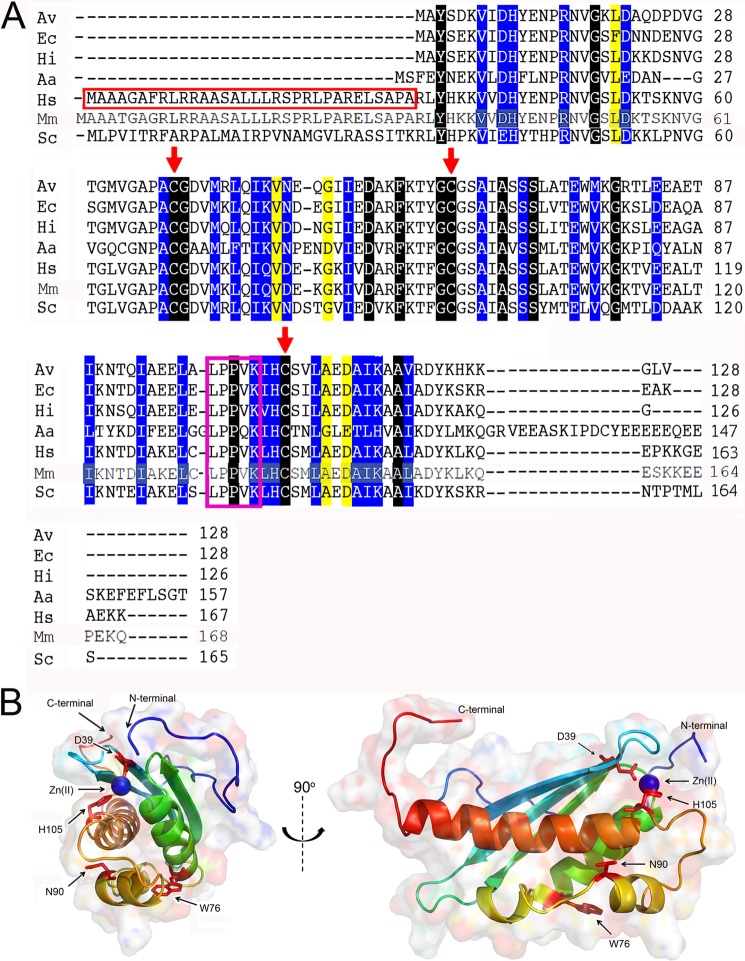
Alignment of sequences of IscU homologues and structural representation of ISCU. A, alignment of sequences of IscU homologues. Analysis (15) of a much larger set of aligned sequences than those represented here showed that the residues highlighted in black were identically conserved, those in blue were conserved, and those in yellow were semi-conserved. The conserved cysteine residues are marked with red arrows, and the conserved LPPVK motif recognized by chaperone proteins is boxed in magenta. Mitochondrial proteins contain an N-terminal sequence that targets ISCU to cross the inner mitochondrial membrane. Excluding this region (boxed in red), human ISCU and E. coli IscU share 77% sequence identity. The numbering systems for mature ISCU and E. coli IscU are identical. Abbreviations used are: Av, A. vinelandii; Ec, Escherichia coli; Hi, H. influenzae; Aa, A. aeolicus; Hs, Homo sapiens; Mm, M. musculus; Sc, S. cerevisiae. B, solution structure of Zn2+ bound M. musculus ISCU (PDB code 1WFZ) (47), which shares ∼98% sequence identity with human ISCU, provides insights into the structure of human ISCU. Similar to other IscU homologues, M. musculus ISCU consists of three β-strands and four α-helices. Residues mutated in this study (Asp-39, Asn-90, and His-105) are shown in red stick format. Asp-39 and His-105 are close to the Zn2+ binding site, and Asn-90 is a solvent-exposed polar residue located in a hydrophobic region. All three residues are highly conserved among IscU homologues, and amino acid substitutions at these sites in E. coli IscU have been shown to perturb the position of the D ⇄ S conformational equilibrium (35). Another important residue Trp-76 is shown in red stick format.