FIGURE 1.
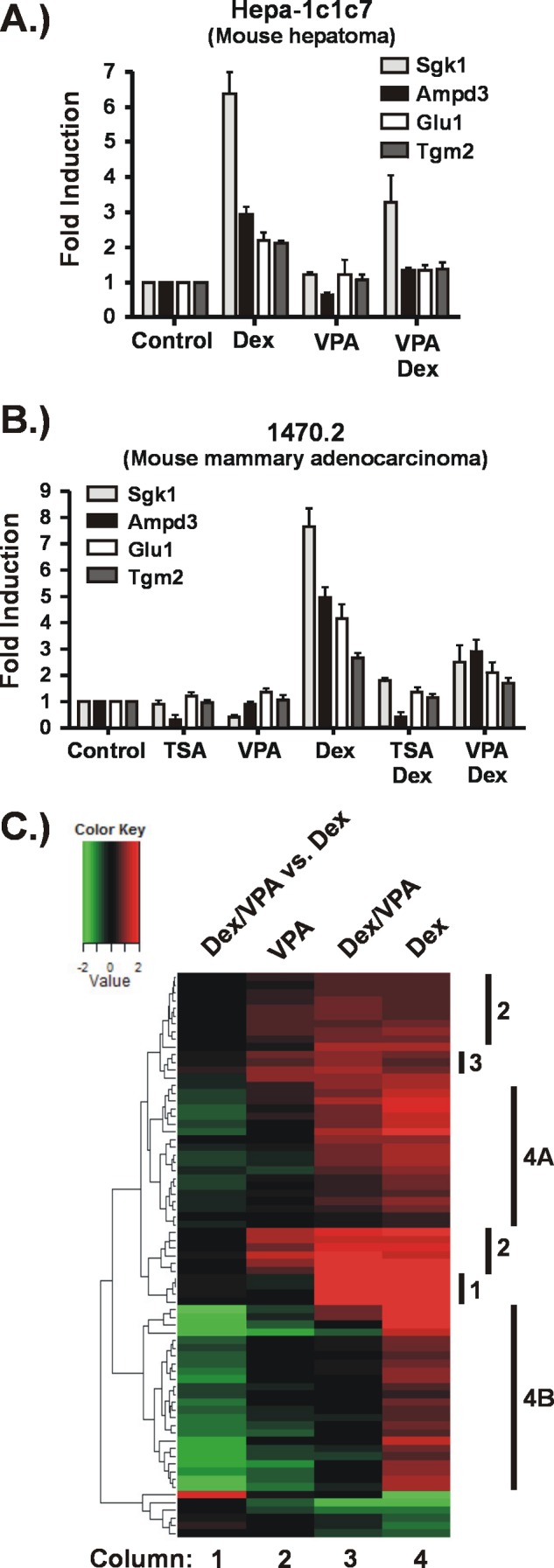
KDACis impair Dex-induced expression of GR target genes. Hepa-1c1c7 cells (A) or 1470.2 cells (B) were exposed to KDACis (VPA (5 mm) or TSA (200 nm)) for 5 h or to Dex (100 nm) for 4 h. For the combination treatments, the KDACis were added to the cells 1 h prior to addition of Dex, which continued for 4 h. A and B, analysis of selected GR target genes by RT-qPCR. The results are represented as -fold inductions relative to the control (untreated). C, effect of VPA on the GR-regulated transcriptome. Cellular RNA was processed for hybridization to microarrays (Affymetrix Mouse GeneChip ST 1.0). Genes whose expression was significantly changed relative to untreated cells were identified for each treatment condition as described under “Experimental Procedures.” The heat map shows only the genes significantly changed by Dex treatment over three biological replicates. Columns 2–4 represent -fold change relative to untreated cells. Column 1 represents the -fold change of the combination treatment compared with Dex alone, indicating the impact of VPA on Dex-regulated gene expression. Denoted to the right of the heat map are groups of genes (1–3, 4A, and 4B) that show similar expression patterns in response to the various treatments and are described under “Results.”
