FIGURE 1.
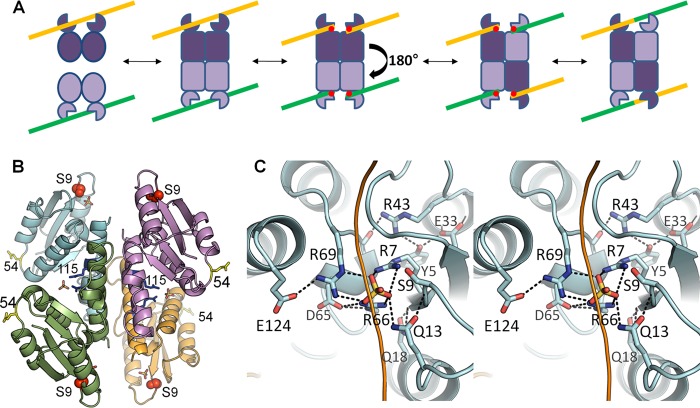
Recombinase mechanism and active site. A, simplified serine recombinase reaction scheme. The recombinase generally binds crossover site DNA as a dimer (yellow and green lines). Two DNA-bound dimers then assemble into a catalytically competent tetramer. Cleavage occurs, forming a transient 5′ phosphoserine linkage (red dots) followed by a putative 180° subunit rotation, which aligns the cleaved DNA ends for religation, yielding a recombinant product. Although WT resolvases usually require accessory factors to trigger tetramerization, the pseudo-WT Sin used here (Q115R/R54E) does not. B, structure of the tetrameric catalytic domain of Sin Q115R/R54E (Protein Data Bank ID code 3PKZ) (16). The nucleophilic serine is shown as red spheres, and bound sulfate ions as sticks. Side chains are also shown for the activating mutation (Q115R, in blue) and the solubilizing mutation (R54E, in yellow). C, active site of Sin. Stereo view shows the Sin active site from the structure in B, in which all of the conserved residues implicated in catalysis form a hydrogen-bonded network surrounding a sulfate ion that lies in the scissile phosphate group-binding pocket (16). The path of a modeled DNA backbone is sketched as a yellow line.