FIGURE 2.
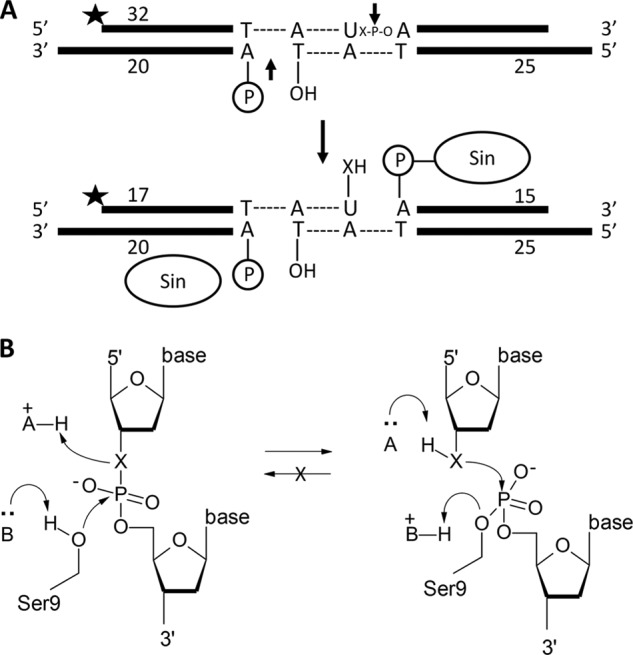
Cleavage assay. A, substrate used in all DNA cleavage assays. Two Sin protomers can bind this crossover site. The top strand (32 nucleotides) is 5′ 32P-labeled, and the scissile position (arrow) contains either a 3′ oxygen or 3′ sulfur (X). The unlabeled bottom strand is nicked at the position of cleavage. Cleavage of the top strand forms a 5′ phosphoserine linkage and a free 3′OH or 3′SH. B, reaction diagram. This represents a hypothetical catalytic mechanism for either the 3′O or 3′S substrate (schematized as X). A general base deprotonates the nucleophilic Ser-9 whereas a general acid protonates the 3′ leaving group. The nick on the opposite strand prevents the reverse (religation) reaction, rendering it a suicide substrate regardless of the identity of X.
