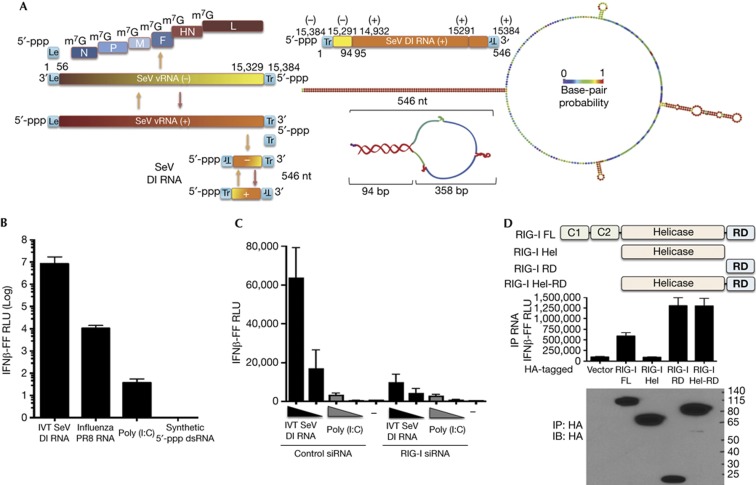
Figure 1.
SeV DI RNA is a potent RIG-I-dependent inducer of IFN-I. (A) Graphic illustrates different RNAs produced during infection of SeV. SeV DI produced from the anti-genomic positive sense (+) RNA consists of both negative and positive sense sequences of the genomic and anti-genomic RNAs resulting in a copy-back structure. The right panel shows the 546-nt SeV DI RNA mapped to the genomic/anti-genomic sequence and a predicted structure of the RNA (RNAfold). Colours on the DI RNA representations indicate base-pair probability (as indicated, red=1; purple=0). (B) 293T-IFNβ-FF-Luc cells were transfected with 50 ng of indicated RNAs and a luciferase assay was performed 24 h later to measure IFNβ promoter-driven luciferase activity. (C) 293T-IFNβ-FF-Luc cells were transfected with control or RIG-I short interfering RNA and 24 h later transfected with 5 and 1 ng of IVT DI RNA and 200 and 40 ng poly (I:C), followed by luciferase assay 24 h later to measure IFNβ promoter-driven luciferase activity. (D) Lysates from 293T cells expressing FL RIG-I or truncated RIG-I Hel, RIG-I C-terminal RD domain or RIG-I Hel-RD were incubated with 0.25 μg of RNA, RIG-I–RNA complexes were immunoprecipitated, and RNA and protein fractions were isolated. RNA was transfected into 293T-IFNβ-FF-Luc cells and 24 h later IFNβ promoter-driven luciferase activity was measured by luciferase assay. Protein fractions were subjected to immunoblotting using HA antibody to assess pull-down efficiency. Data are representative of at least three independent experiments and error bars indicate mean±s.d. DI, defective interfering; FL, full length; Hel, Helicase domain; Hel-RD, Helicase regulatory domain; IFN-I, type-I interferon; IVT, in vitro transcribed; RD, regulatory domain; SeV, Sendai virus.