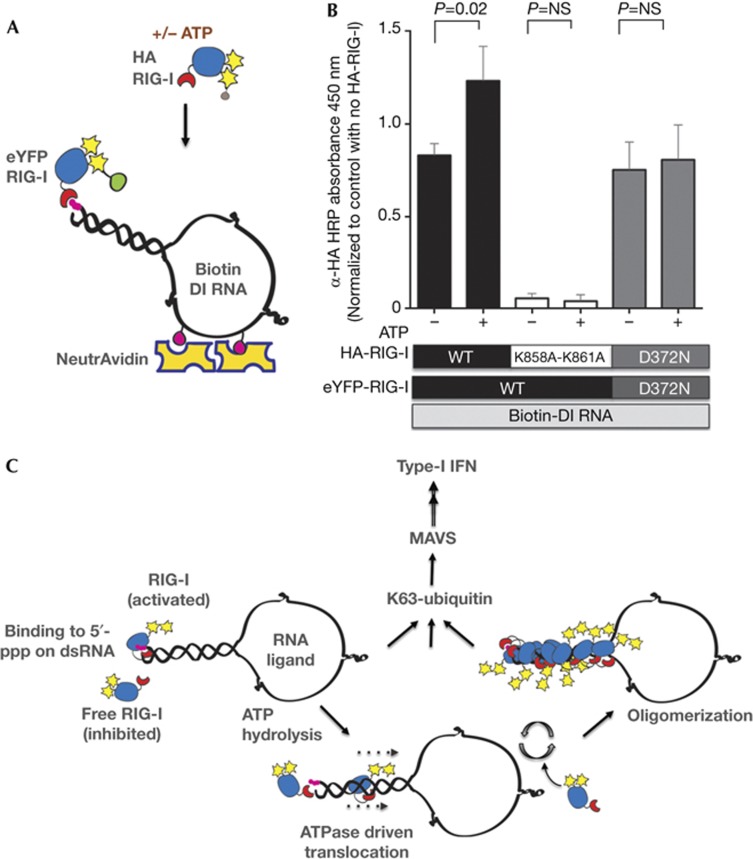
Figure 5.
Mechanism for ATP-driven formation of RIG-I oligomers on RNA. (A) Schematic of the experiment highlighting immobilization of Biotin-labelled RNA onto NeutrAvidin and saturation with eYFP-RIG-I, followed by binding of HA-RIG-I in the presence or absence of ATP. (B) 80 ng of Biotin-labelled DI RNA was immobilized on NeutrAvidin-coated wells, washed and 1 μg of eYFP-RIG-I added (WT or D372N mutant). Following washes, 0.2 μg of HA-RIG-I (WT, K858A-K861A or D372N) was added in the presence or absence of 1 mM ATP as indicated. Bound HA-RIG-I was detected using α-HA-HRP antibody. Data are representative of three independent experiments and error bars indicate mean±s.d. The P-value was calculated using a Student’s unpaired t test. (C) A proposed model of RIG-I activation. Upon binding to 5′-ppp on dsRNA ligand, RIG-I is activated with a conformation change and ATP hydrolysis. Our data suggest that ATP hydrolysis by RNA-bound RIG-I allows exposure of 5′-ppp and recruitment of additional RIG-I molecules on long dsRNA forming RIG-I oligomers. As demonstrated previously, upon K63-linked polyubiquitination or polyubiquitin binding, RIG-I is further activated and binds MAVS to induce IFN-I production. 5′-ppp, 5′-triphosphate; DI, defective interfering; dsRNA, double-stranded RNA; HRP, horseradish peroxidase; IFN-I, type-1 interferon; MAVS, mitochondrial antiviral signal; NS, not significant; WT, wild type.