Table 2. Classification results with different settings.
| Mutation feature | Mutation feature + Personal features | |||||||
|---|---|---|---|---|---|---|---|---|
Number of Hidden Nodes 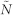
|
Training Time for each fold (seconds) | Testing Time for each fold (seconds) | Training Accuracy (average) | Testing Accuracy (average) | Training Time for each fold (seconds) | Testing Time for each fold (seconds) | Training Accuracy (average) | Testing Accuracy (average) |
| 50 | 0.0144 | 0.0002 | 0.8155 | 0.7917 | 0.0145 | 0.0004 | 0.8004 | 0.8333 |
| 100 | 0.0379 | 0.0004 | 0.8155 | 0.7262 | 0.0516 | 0.0002 | 0.9303 | 0.8810 |
| 150 | 0.0555 | 0.0003 | 0.8155 | 0.7024 | 0.1147 | 0.0003 | 0.9762 | 0.9583 |
| 200 | 0.0767 | 0.0010 | 0.8155 | 0.6845 | 0.1461 | 0.0006 | 0.9762 | 0.9286 |
| 250 | 0.0898 | 0.0006 | 0.8155 | 0.6607 | 0.1649 | 0.0005 | 0.9762 | 0.9226 |
| 300 | 0.1010 | 0.0005 | 0.8156 | 0.6607 | 0.1642 | 0.0005 | 0.9763 | 0.9107 |
| 350 | 0.1076 | 0.0009 | 0.8155 | 0.6607 | 0.1729 | 0.0007 | 0.9762 | 0.9048 |
| 400 | 0.1142 | 0.0010 | 0.8154 | 0.6488 | 0.1827 | 0.0007 | 0.9763 | 0.8869 |
| 450 | 0.1229 | 0.0006 | 0.8156 | 0.6488 | 0.1887 | 0.0009 | 0.9762 | 0.8869 |
| 500 | 0.1323 | 0.0010 | 0.8155 | 0.6488 | 0.1978 | 0.0009 | 0.9763 | 0.8810 |
This table shows the prediction results of the response level to the specific inhibitors for the observed patients. Two feature sets, one including the mutation feature only while the other involving both the mutation feature and personal features, are applied for a comparison. Extreme learning machines and leave-one-out cross validation are used in the calculation. The number of hidden nodes varies from 50 to 500 at a step of 50, and the execution time and accuracy are shown in the table.
