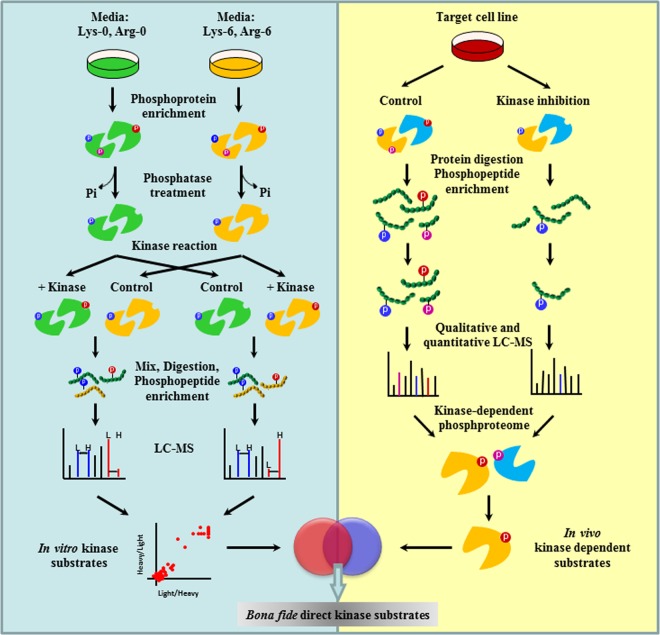
Fig. 1.
Methodology of proKALIP to identify kinase substrates. proKALIP combines in vitro kinase reactions and in vivo phosphoproteomics. In the in vitro kinase reaction, substrate proteins are isolated from cell lysates through affinity purification, dephosphorylated by alkaline phosphatase, and rephosphorylated by kinase of interest. After an in vitro kinase reaction, phosphopeptides are further enriched and analyzed by mass spectrometry for sequencing and site identification. Theoretical substrate has a higher intensity in kinase+ sample compared with the control in SILAC experiments. In in vivo phosphoproteomics, kinase dependent phosphorylation events are identified by comparing two phosphoproteomes with kinase perturbations. Genuine substrates are the phosphopeptides present within both data sets from in vitro kinase reaction and in vivo phosphoproteomics.