Abstract
In the crystal structure of the title compound, C16H24N4O4, molecules are linked by N—H⋯O hydrogen bonds between the carbonyl groups of the carbamoyl and amido functional groups and the amino groups, and by N—H⋯N hydrogen bonds between the amino group and the pyridine ring, forming two-dimensional networks parallel to the ab plane.
Related literature
For the synthesis, properties and biological activity of 2-hydrazinopyridine derivatives, see: Ardisson et al. (2005 ▶); Jurisson & Lydon (1999 ▶); Abrams et al. (1994 ▶); Liu et al. (2011 ▶); Lu et al. (2011 ▶); Schwartz et al. (1990 ▶). For the crystal structures of related compounds, see: Banerjee et al. (2005 ▶); Rose et al. (1998 ▶); Zora et al. (2006 ▶). For synthesis, see: Cugola et al. (1995 ▶).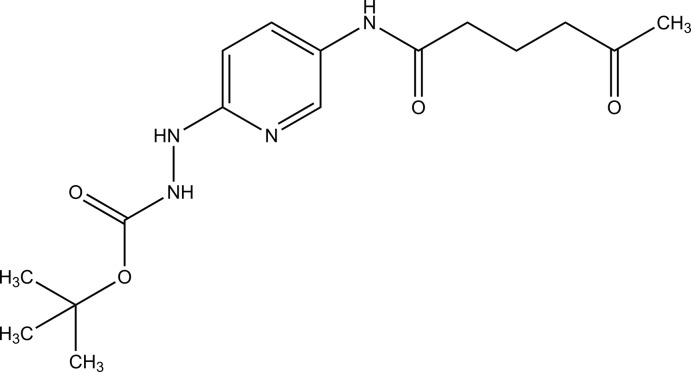
Experimental
Crystal data
C16H24N4O4
M r = 336.39
Triclinic,

a = 6.2598 (4) Å
b = 9.2822 (6) Å
c = 16.0437 (12) Å
α = 84.387 (6)°
β = 88.957 (6)°
γ = 79.358 (6)°
V = 911.79 (11) Å3
Z = 2
Mo Kα radiation
μ = 0.09 mm−1
T = 293 K
0.84 × 0.17 × 0.06 mm
Data collection
Bruker–Nonius KappaCCD diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.928, T max = 0.994
20299 measured reflections
3295 independent reflections
2187 reflections with I > 2σ(I)
R int = 0.038
Refinement
R[F 2 > 2σ(F 2)] = 0.053
wR(F 2) = 0.147
S = 1.04
3295 reflections
221 parameters
H-atom parameters constrained
Δρmax = 0.29 e Å−3
Δρmin = −0.25 e Å−3
Data collection: COLLECT (Nonius, 1998 ▶); cell refinement: DIRAX/LSQ (Duisenberg, 1992 ▶); data reduction: EVALCCD (Duisenberg et al., 2003 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: OLEX2 (Dolomanov et al., 2009 ▶); software used to prepare material for publication: publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S1600536813024598/ff2115sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813024598/ff2115Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813024598/ff2115Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O3i | 0.86 | 2.06 | 2.888 (2) | 161 |
| N3—H3⋯N2ii | 0.86 | 2.21 | 2.957 (3) | 145 |
| N4—H4⋯O2iii | 0.86 | 2.06 | 2.827 (3) | 149 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Acknowledgments
This publication was supported by a grant from the Ligue Nationale contre le Cancer (Comité Aquitaine-Charentes, Bordeaux, France).
supplementary crystallographic information
1. Comment
Radioisotopes conjugated to proteins provide a means for imaging and treatment of disease. The bifunctional 2-hydrazinopyridine derivatives are useful linker molecules for attaching metal ions such as 99mTc to macromolecules (Ardisson et al., 2005; Jurisson & Lydon, 1999). Hence, this 2-hydrazinopyridinyl moiety has previously been used for labeling bioactive molecules (Abrams et al., 1994; Banerjee et al. 2005; Rose et al., 1998; Schwartz et al., 1990). Thus, the use of Tc-labeled hydrazine derivatives continues to undergo further development (Liu et al., 2011; Lu et al., 2011). The wide spectrum of medicinal applications of this class of radiolabeled chelates prompted us to work in this domain and we report herein on the synthesis and crystal structure of the title compound, designed as a potential chelate for 99mTc.
The title compound, C16H24N4O4, has the triclinic (P1) symmetry. It crystalizes with one molecule in the asymmetric unit. In the crystal, the molecules are linked together by N—H···O hydrogen bonding between the carbonyl groups of the carbamoyl and amido functional groups and the amino groups and by N—H···N hydrogen bonding between amino and pyridine moiety leading to a two-dimensional network within the ab plane. The network cohesion in the 3rd direction is assured by Van der Waals interactions and H-bond like interactions between the carbonyl and the BOC group.
2. Experimental
To a stirred solution of 5-oxohexanoic acid (2.45 mmol) in 15 ml of tetrahydrofurane was added triethylamine (2.45 mmol). After 10 min of stirring at room temperature was added isobutyl chloroformate (2.45 mmol). The reaction mixture was stirred at room temperature for 5 h, then 2-(t-butoxycarbonyl hydrazine)-5-amino-pyridine (2.23 mmol) (Cugola et al.,19955) was added and the reaction stirred for 12 h. The mixture was evaporated to dryness, and the residue was triturated in water. The solid precipitate was filtered off and washed with water then with ethanol, and purified by column chromatography using CHCl3/methanol (9/1, v/v) as eluent to give the title compound as white crystals (Rf = 1/4). Yield is 48%. The single-crystal of the title product was obtained by slow crystallization from a mixture DMSO/methanol (9/1, v/v). M.p. = 219°C. IR (KBr), ν/cm-1: 3270, 3230, 3185, 3082, 2982, 1704, 1662, 1601, 1537, 1276, 1182. 1H NMR (300 MHz, DMSO-d6, 298 K): δ = 1.41 (s, 9H, 3 CH3), 1.75 (qt, 2H, J = 6.10, CH2), 2.09 (s, 3H, CH3), 2.26 (t, 2H, J = 6.10, CH2), 2.48 (t, 2H, J = 6.10, CH2), 6.49 (d, 1H, J = 7.50, H-3), 7.71 (dd, 1H, J = 7.50 and 1.55, H-4), 7.95 (s, 1H, NH), 8.20 (d, 1H, J = 1.55, H-6), 8.75 (s, 1H, NH), 9.70 (s, 1H, NH). Anal. Calcd. for C16H24N4O4: C, 57.13; H, 7.19; N, 16.66 Found: C, 57.26; H, 7.25; N, 16.52.
3. Refinement
Crystallographic data were collected at 293 K on a Brucker nonius k-CCD diffractometer with monochromatic Mo—Kα radiation (λ = 0.71073 Å). At 293 K, the full sphere data collection was performed using φ scans and ω scans. The unit cell determination and data reduction were performed using DIRAX/LSQ (Duisenberg, 1992) and Collect (Nonius, 1998) programs on the full set of data. The crystal structure was solved by direct methods and successive Fourier difference syntheses with SHELXS97 program (Sheldrick, 2008). The refinements of the crystal structure were performed on F2 by weighted anisotropic full-matrix least squares methods using the SHELXL97 program (Sheldrick, 2008). Both pieces of software were used within OLEX2 package (Dolomanov et al., 2009). All the non-H atoms were refined with anisotropic temperature parameters. The positions of the H atoms were deduced from coordinates of the non-H atoms and confirmed by Fourier synthesis and treated according to the riding model during refinement with isotropic displacement parameters, corresponding to the atom they are linked to. H atoms were included for structure factor calculations but not refined.
Figures
Fig. 1.
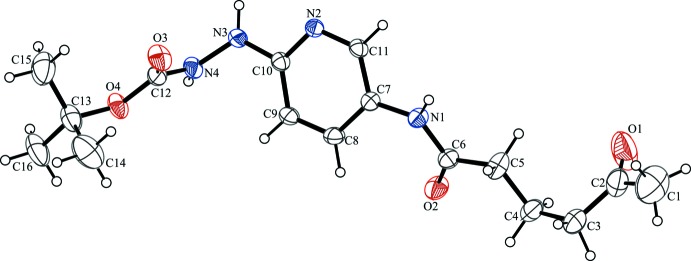
The molecular structure of the title compound. Displacement ellipsoids are drawn at the 30% probability level.
Fig. 2.

Crystal packing of the title compound viewed down the a axis. Only hydrogen atoms involved in hydrogen bonding (dashed lines) are shown.
Crystal data
| C16H24N4O4 | Z = 2 |
| Mr = 336.39 | F(000) = 360 |
| Triclinic, P1 | Dx = 1.225 Mg m−3 |
| a = 6.2598 (4) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 9.2822 (6) Å | Cell parameters from 8003 reflections |
| c = 16.0437 (12) Å | θ = 3.2–25.3° |
| α = 84.387 (6)° | µ = 0.09 mm−1 |
| β = 88.957 (6)° | T = 293 K |
| γ = 79.358 (6)° | Plate, colourless |
| V = 911.79 (11) Å3 | 0.84 × 0.17 × 0.06 mm |
Data collection
| Bruker–Nonius KappaCCD diffractometer | 2187 reflections with I > 2σ(I) |
| intensities from φ scan and ω scan | Rint = 0.038 |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | θmax = 25.3°, θmin = 3.2° |
| Tmin = 0.928, Tmax = 0.994 | h = −7→7 |
| 20299 measured reflections | k = −11→11 |
| 3295 independent reflections | l = −19→19 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.053 | H-atom parameters constrained |
| wR(F2) = 0.147 | w = 1/[σ2(Fo2) + (0.061P)2 + 0.4578P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.04 | (Δ/σ)max = 0.001 |
| 3295 reflections | Δρmax = 0.29 e Å−3 |
| 221 parameters | Δρmin = −0.25 e Å−3 |
| 0 restraints |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N2 | 0.2722 (3) | 0.91380 (18) | 0.96382 (11) | 0.0383 (4) | |
| N1 | 0.8167 (3) | 0.79747 (19) | 0.88090 (12) | 0.0420 (5) | |
| H1 | 0.8436 | 0.8828 | 0.8633 | 0.050* | |
| C8 | 0.6222 (4) | 0.6875 (2) | 0.99807 (14) | 0.0452 (6) | |
| H8 | 0.7405 | 0.6120 | 1.0105 | 0.054* | |
| N3 | 0.0661 (3) | 0.8185 (2) | 1.06579 (12) | 0.0480 (5) | |
| H3 | −0.0492 | 0.8704 | 1.0424 | 0.058* | |
| O4 | 0.0851 (3) | 0.71283 (17) | 1.28015 (10) | 0.0579 (5) | |
| C10 | 0.2624 (3) | 0.8083 (2) | 1.02536 (13) | 0.0376 (5) | |
| C11 | 0.4549 (3) | 0.9045 (2) | 0.91869 (13) | 0.0388 (5) | |
| H11 | 0.4618 | 0.9774 | 0.8752 | 0.047* | |
| O2 | 0.9342 (3) | 0.55429 (18) | 0.87934 (12) | 0.0663 (5) | |
| N4 | 0.0544 (3) | 0.7449 (2) | 1.14426 (12) | 0.0476 (5) | |
| H4 | 0.0149 | 0.6605 | 1.1495 | 0.057* | |
| C7 | 0.6329 (3) | 0.7943 (2) | 0.93254 (13) | 0.0369 (5) | |
| O3 | 0.1590 (3) | 0.92288 (18) | 1.21135 (11) | 0.0640 (5) | |
| C6 | 0.9535 (4) | 0.6804 (2) | 0.85656 (14) | 0.0439 (6) | |
| C9 | 0.4362 (4) | 0.6941 (2) | 1.04441 (14) | 0.0462 (6) | |
| H9 | 0.4260 | 0.6226 | 1.0884 | 0.055* | |
| C12 | 0.1045 (4) | 0.8044 (2) | 1.21198 (15) | 0.0461 (6) | |
| C5 | 1.1308 (4) | 0.7170 (3) | 0.79916 (18) | 0.0620 (7) | |
| H5A | 1.0644 | 0.7759 | 0.7499 | 0.074* | |
| H5B | 1.2121 | 0.7776 | 0.8272 | 0.074* | |
| C2 | 1.4473 (6) | 0.7179 (4) | 0.6461 (2) | 0.0740 (8) | |
| C13 | 0.1179 (5) | 0.7546 (3) | 1.36427 (16) | 0.0663 (8) | |
| C4 | 1.2840 (5) | 0.5918 (4) | 0.7717 (2) | 0.0931 (12) | |
| H4A | 1.2109 | 0.5423 | 0.7334 | 0.112* | |
| H4B | 1.3307 | 0.5222 | 0.8199 | 0.112* | |
| O1 | 1.2875 (5) | 0.7200 (4) | 0.60720 (16) | 0.1353 (12) | |
| C15 | −0.0383 (7) | 0.8914 (4) | 1.3805 (2) | 0.1030 (13) | |
| H15A | 0.0014 | 0.9742 | 1.3473 | 0.154* | |
| H15B | −0.0341 | 0.9063 | 1.4388 | 0.154* | |
| H15C | −0.1825 | 0.8815 | 1.3658 | 0.154* | |
| C3 | 1.4820 (5) | 0.6376 (5) | 0.7287 (2) | 0.1002 (13) | |
| H3A | 1.5420 | 0.6983 | 0.7646 | 0.120* | |
| H3B | 1.5906 | 0.5497 | 0.7237 | 0.120* | |
| C1 | 1.6198 (8) | 0.7953 (5) | 0.6113 (3) | 0.1350 (18) | |
| H1A | 1.6100 | 0.8071 | 0.5513 | 0.203* | |
| H1B | 1.7593 | 0.7387 | 0.6279 | 0.203* | |
| H1C | 1.6022 | 0.8903 | 0.6321 | 0.203* | |
| C16 | 0.0675 (7) | 0.6234 (4) | 1.41952 (19) | 0.0943 (11) | |
| H16A | −0.0786 | 0.6116 | 1.4092 | 0.142* | |
| H16B | 0.0821 | 0.6391 | 1.4772 | 0.142* | |
| H16C | 0.1669 | 0.5363 | 1.4072 | 0.142* | |
| C14 | 0.3522 (7) | 0.7691 (5) | 1.3740 (2) | 0.1111 (13) | |
| H14A | 0.4454 | 0.6793 | 1.3617 | 0.167* | |
| H14B | 0.3770 | 0.7881 | 1.4304 | 0.167* | |
| H14C | 0.3832 | 0.8491 | 1.3359 | 0.167* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N2 | 0.0429 (11) | 0.0323 (10) | 0.0384 (10) | −0.0052 (8) | 0.0023 (8) | −0.0001 (8) |
| N1 | 0.0428 (11) | 0.0303 (10) | 0.0531 (12) | −0.0092 (8) | 0.0086 (9) | −0.0018 (8) |
| C8 | 0.0449 (14) | 0.0346 (12) | 0.0516 (14) | 0.0005 (10) | 0.0001 (11) | 0.0037 (10) |
| N3 | 0.0438 (12) | 0.0504 (12) | 0.0432 (11) | −0.0003 (9) | 0.0058 (9) | 0.0115 (9) |
| O4 | 0.0911 (13) | 0.0425 (10) | 0.0428 (10) | −0.0241 (9) | 0.0090 (8) | 0.0042 (7) |
| C10 | 0.0424 (13) | 0.0334 (12) | 0.0373 (12) | −0.0090 (9) | 0.0000 (9) | −0.0020 (9) |
| C11 | 0.0465 (14) | 0.0312 (12) | 0.0383 (12) | −0.0090 (10) | 0.0011 (10) | 0.0016 (9) |
| O2 | 0.0826 (13) | 0.0343 (10) | 0.0792 (13) | −0.0052 (9) | 0.0231 (10) | −0.0060 (9) |
| N4 | 0.0630 (13) | 0.0354 (10) | 0.0437 (11) | −0.0130 (9) | 0.0095 (9) | 0.0049 (9) |
| C7 | 0.0384 (12) | 0.0313 (11) | 0.0424 (12) | −0.0096 (9) | 0.0017 (9) | −0.0046 (9) |
| O3 | 0.0948 (14) | 0.0404 (10) | 0.0608 (11) | −0.0279 (9) | 0.0081 (10) | 0.0031 (8) |
| C6 | 0.0468 (14) | 0.0367 (13) | 0.0488 (14) | −0.0076 (10) | 0.0007 (10) | −0.0077 (10) |
| C9 | 0.0507 (14) | 0.0358 (13) | 0.0471 (13) | −0.0022 (10) | 0.0045 (11) | 0.0100 (10) |
| C12 | 0.0535 (15) | 0.0349 (13) | 0.0479 (14) | −0.0088 (11) | 0.0110 (11) | 0.0050 (11) |
| C5 | 0.0559 (16) | 0.0620 (17) | 0.0733 (18) | −0.0168 (13) | 0.0195 (13) | −0.0244 (14) |
| C2 | 0.073 (2) | 0.089 (2) | 0.0620 (19) | −0.0139 (17) | 0.0123 (17) | −0.0204 (17) |
| C13 | 0.096 (2) | 0.0598 (18) | 0.0455 (15) | −0.0248 (15) | 0.0073 (14) | 0.0013 (13) |
| C4 | 0.089 (2) | 0.079 (2) | 0.087 (2) | 0.0318 (18) | 0.0401 (19) | 0.0168 (18) |
| O1 | 0.112 (2) | 0.239 (4) | 0.0606 (16) | −0.050 (2) | −0.0055 (15) | −0.0106 (19) |
| C15 | 0.154 (4) | 0.075 (2) | 0.080 (2) | −0.018 (2) | 0.038 (2) | −0.0189 (18) |
| C3 | 0.063 (2) | 0.147 (3) | 0.071 (2) | 0.027 (2) | 0.0151 (16) | 0.000 (2) |
| C1 | 0.126 (4) | 0.117 (4) | 0.169 (5) | −0.046 (3) | 0.046 (3) | −0.009 (3) |
| C16 | 0.156 (3) | 0.077 (2) | 0.0518 (18) | −0.037 (2) | 0.0132 (19) | 0.0112 (16) |
| C14 | 0.122 (3) | 0.133 (4) | 0.086 (3) | −0.050 (3) | −0.025 (2) | 0.011 (2) |
Geometric parameters (Å, º)
| N2—C10 | 1.330 (3) | C5—C4 | 1.462 (4) |
| N2—C11 | 1.336 (3) | C2—O1 | 1.185 (4) |
| N1—H1 | 0.8600 | C2—C3 | 1.454 (4) |
| N1—C7 | 1.409 (3) | C2—C1 | 1.476 (5) |
| N1—C6 | 1.339 (3) | C13—C15 | 1.494 (4) |
| C8—H8 | 0.9300 | C13—C16 | 1.513 (4) |
| C8—C7 | 1.382 (3) | C13—C14 | 1.510 (5) |
| C8—C9 | 1.365 (3) | C4—H4A | 0.9700 |
| N3—H3 | 0.8600 | C4—H4B | 0.9700 |
| N3—C10 | 1.372 (3) | C4—C3 | 1.517 (4) |
| N3—N4 | 1.380 (2) | C15—H15A | 0.9600 |
| O4—C12 | 1.336 (3) | C15—H15B | 0.9600 |
| O4—C13 | 1.468 (3) | C15—H15C | 0.9600 |
| C10—C9 | 1.386 (3) | C3—H3A | 0.9700 |
| C11—H11 | 0.9300 | C3—H3B | 0.9700 |
| C11—C7 | 1.372 (3) | C1—H1A | 0.9600 |
| O2—C6 | 1.219 (3) | C1—H1B | 0.9600 |
| N4—H4 | 0.8600 | C1—H1C | 0.9600 |
| N4—C12 | 1.333 (3) | C16—H16A | 0.9600 |
| O3—C12 | 1.209 (3) | C16—H16B | 0.9600 |
| C6—C5 | 1.495 (3) | C16—H16C | 0.9600 |
| C9—H9 | 0.9300 | C14—H14A | 0.9600 |
| C5—H5A | 0.9700 | C14—H14B | 0.9600 |
| C5—H5B | 0.9700 | C14—H14C | 0.9600 |
| C10—N2—C11 | 117.63 (18) | O4—C13—C16 | 101.9 (2) |
| C7—N1—H1 | 116.9 | O4—C13—C14 | 109.3 (3) |
| C6—N1—H1 | 116.9 | C15—C13—C16 | 110.7 (3) |
| C6—N1—C7 | 126.27 (18) | C15—C13—C14 | 112.9 (3) |
| C7—C8—H8 | 120.4 | C14—C13—C16 | 110.8 (3) |
| C9—C8—H8 | 120.4 | C5—C4—H4A | 109.1 |
| C9—C8—C7 | 119.3 (2) | C5—C4—H4B | 109.1 |
| C10—N3—H3 | 120.2 | C5—C4—C3 | 112.4 (3) |
| C10—N3—N4 | 119.59 (18) | H4A—C4—H4B | 107.8 |
| N4—N3—H3 | 120.2 | C3—C4—H4A | 109.1 |
| C12—O4—C13 | 121.02 (19) | C3—C4—H4B | 109.1 |
| N2—C10—N3 | 114.92 (18) | C13—C15—H15A | 109.5 |
| N2—C10—C9 | 122.0 (2) | C13—C15—H15B | 109.5 |
| N3—C10—C9 | 123.07 (19) | C13—C15—H15C | 109.5 |
| N2—C11—H11 | 117.9 | H15A—C15—H15B | 109.5 |
| N2—C11—C7 | 124.12 (19) | H15A—C15—H15C | 109.5 |
| C7—C11—H11 | 117.9 | H15B—C15—H15C | 109.5 |
| N3—N4—H4 | 120.0 | C2—C3—C4 | 116.5 (3) |
| C12—N4—N3 | 120.07 (19) | C2—C3—H3A | 108.2 |
| C12—N4—H4 | 120.0 | C2—C3—H3B | 108.2 |
| C8—C7—N1 | 123.77 (19) | C4—C3—H3A | 108.2 |
| C11—C7—N1 | 118.68 (19) | C4—C3—H3B | 108.2 |
| C11—C7—C8 | 117.5 (2) | H3A—C3—H3B | 107.3 |
| N1—C6—C5 | 114.7 (2) | C2—C1—H1A | 109.5 |
| O2—C6—N1 | 122.5 (2) | C2—C1—H1B | 109.5 |
| O2—C6—C5 | 122.8 (2) | C2—C1—H1C | 109.5 |
| C8—C9—C10 | 119.5 (2) | H1A—C1—H1B | 109.5 |
| C8—C9—H9 | 120.3 | H1A—C1—H1C | 109.5 |
| C10—C9—H9 | 120.3 | H1B—C1—H1C | 109.5 |
| N4—C12—O4 | 109.39 (19) | C13—C16—H16A | 109.5 |
| O3—C12—O4 | 125.6 (2) | C13—C16—H16B | 109.5 |
| O3—C12—N4 | 125.0 (2) | C13—C16—H16C | 109.5 |
| C6—C5—H5A | 108.3 | H16A—C16—H16B | 109.5 |
| C6—C5—H5B | 108.3 | H16A—C16—H16C | 109.5 |
| H5A—C5—H5B | 107.4 | H16B—C16—H16C | 109.5 |
| C4—C5—C6 | 116.1 (2) | C13—C14—H14A | 109.5 |
| C4—C5—H5A | 108.3 | C13—C14—H14B | 109.5 |
| C4—C5—H5B | 108.3 | C13—C14—H14C | 109.5 |
| O1—C2—C3 | 121.4 (3) | H14A—C14—H14B | 109.5 |
| O1—C2—C1 | 120.7 (4) | H14A—C14—H14C | 109.5 |
| C3—C2—C1 | 117.8 (4) | H14B—C14—H14C | 109.5 |
| O4—C13—C15 | 110.7 (3) | ||
| N2—C10—C9—C8 | −1.1 (3) | C7—N1—C6—C5 | 178.0 (2) |
| N2—C11—C7—N1 | −179.53 (19) | C7—C8—C9—C10 | −0.6 (4) |
| N2—C11—C7—C8 | −0.9 (3) | C6—N1—C7—C8 | 34.9 (3) |
| N1—C6—C5—C4 | 179.6 (3) | C6—N1—C7—C11 | −146.5 (2) |
| N3—C10—C9—C8 | 176.3 (2) | C6—C5—C4—C3 | −168.4 (3) |
| N3—N4—C12—O4 | −178.94 (18) | C9—C8—C7—N1 | −179.9 (2) |
| N3—N4—C12—O3 | 1.0 (4) | C9—C8—C7—C11 | 1.5 (3) |
| C10—N2—C11—C7 | −0.7 (3) | C12—O4—C13—C15 | 59.1 (3) |
| C10—N3—N4—C12 | 82.6 (3) | C12—O4—C13—C16 | 176.9 (2) |
| C11—N2—C10—N3 | −175.89 (18) | C12—O4—C13—C14 | −65.8 (3) |
| C11—N2—C10—C9 | 1.7 (3) | C5—C4—C3—C2 | −70.8 (4) |
| O2—C6—C5—C4 | −0.5 (4) | C13—O4—C12—N4 | −176.3 (2) |
| N4—N3—C10—N2 | −160.78 (19) | C13—O4—C12—O3 | 3.8 (4) |
| N4—N3—C10—C9 | 21.6 (3) | O1—C2—C3—C4 | −15.7 (5) |
| C7—N1—C6—O2 | −1.8 (4) | C1—C2—C3—C4 | 165.3 (3) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O3i | 0.86 | 2.06 | 2.888 (2) | 161 |
| N3—H3···N2ii | 0.86 | 2.21 | 2.957 (3) | 145 |
| N4—H4···O2iii | 0.86 | 2.06 | 2.827 (3) | 149 |
Symmetry codes: (i) −x+1, −y+2, −z+2; (ii) −x, −y+2, −z+2; (iii) −x+1, −y+1, −z+2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: FF2115).
References
- Abrams, M. J., Bridger, G. J., Schwartz, D. A., Padmanabhan, S. & Ultee, M. E. (1994). World Patent WO 94/10149.
- Ardisson, V., Mathieu, J. P., Ghezzi, C. & Fagret, D. (2005). Med. Nucl. 29, 168–178.
- Banerjee, S. R., Schaffer, P., Babich, J. W., Valliant, J. F. & Zubieta, J. (2005). Dalton Trans. pp. 3886–3897. [DOI] [PubMed]
- Cugola, A., Di Fabio, R. & Pentasuglia, G. (1995). World Patent WO 95/10517.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Duisenberg, A. J. M. (1992). J. Appl. Cryst. 25, 92–96.
- Duisenberg, A. J. M., Kroon-Batenburg, L. M. J. & Schreurs, A. M. M. (2003). J. Appl. Cryst. 36, 220–229.
- Jurisson, S. S. & Lydon, J. D. (1999). Chem. Rev. 99, 2205–2218. [DOI] [PubMed]
- Liu, L., Zhang, M., Zhong, G. & Wang, X. (2011). J. Radioanal. Nucl. Chem. 287, 847–852.
- Lu, J., Pang, Y., Xie, F., Guo, H.-J., Li, Y., Yang, Z. & Wang, X.-B. (2011). Nucl. Med. Biol. 38, 557–565. [DOI] [PubMed]
- Nonius (1998). COLLECT Nonius BV, Delft, The Netherlands.
- Rose, D. J., Maresca, K. P., Nicholson, T., Davison, A., Jones, A. G., Babich, J., Fischman, A., Graham, W., DeBord, J. R. D. & Zubieta, J. (1998). Inorg. Chem. 37, 2701–2716. [DOI] [PubMed]
- Schwartz, D. A., Abrams, M. J., Giandomenico, C. M. & Zubieta, J. A. (1990). Eur. Patent EP 384769.
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Zora, M., Turgut, G., Odabaşoğlu, M. & Büyükgüngör, O. (2006). Acta Cryst. E62, o2677–o2679.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S1600536813024598/ff2115sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813024598/ff2115Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813024598/ff2115Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report


