Figure 1.
Loss of NEK8 leads to accumulation of DNA damage.
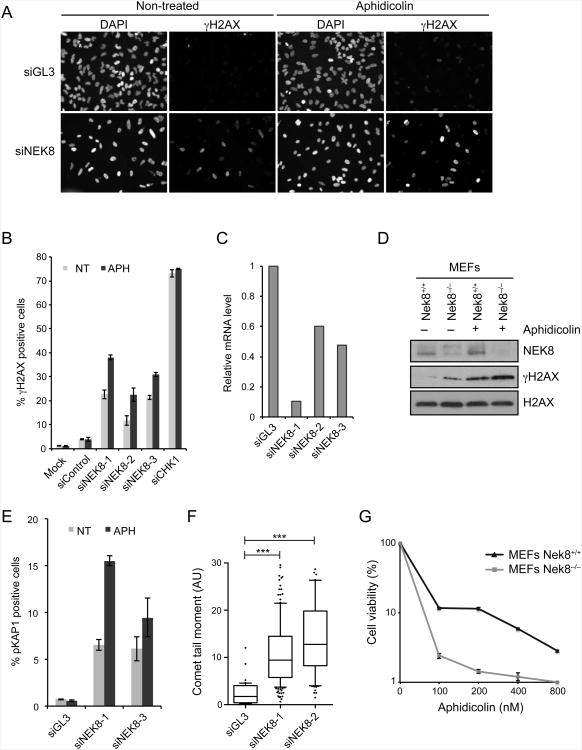
(A-B) Induction of γH2AX upon NEK8 knockdown. HeLa cells were transfected with siRNAs targeting luciferase (siGL3) or NEK8 (siNEK8), treated with or without aphidicolin (APH, 400 nM) for 24 h, stained with γH2AX antibodies and DAPI, and analyzed by immunofluorescence. (A) Representative images of γH2AX and DAPI staining. (B) Quantification of percent γH2AX-positive cells. Each data point represents the mean of three experimental replicates. Error bars represent standard error of mean (SEM, n=3). (C) mRNA levels of NEK8 were assessed by RT-qPCR. (D) γH2AX levels in wild-type (Nek8+/+) and knockout (Nek8−/−) MEFs determined by western blotting. APH (400 nM) was used for 18 h. γtubulin was used as a loading control. (E) Quantification of percent pKAP1-positive cells. Each data point represents the mean of three experimental replicates. Error bars represent standard error of mean (SEM, n=3). (F) Quantification of tail moment from neutral comet assay in HeLa cells transfected with siGL3 or siNEK8. Box and whiskers indicate 25-75 and 10-90 percentiles, respectively. The lines represent the median values. Asterisks indicate the P-value of the statistical test (Mann-Whitney rank sum t-test, ***p < 0.0001). Data not included between the whiskers are plotted as outliers (dots). (G) Sensitivity of Nek8−/− MEFs to replication stress. Cells were treated with increasing doses of aphidicolin for 18 h. Surviving colonies were scored 12 days later. Each data point represents the mean of three experimental replicates. Error bars represent standard error of mean (SEM, n=3). In all panels, the data are from three independent experiments. See also Figure S1.