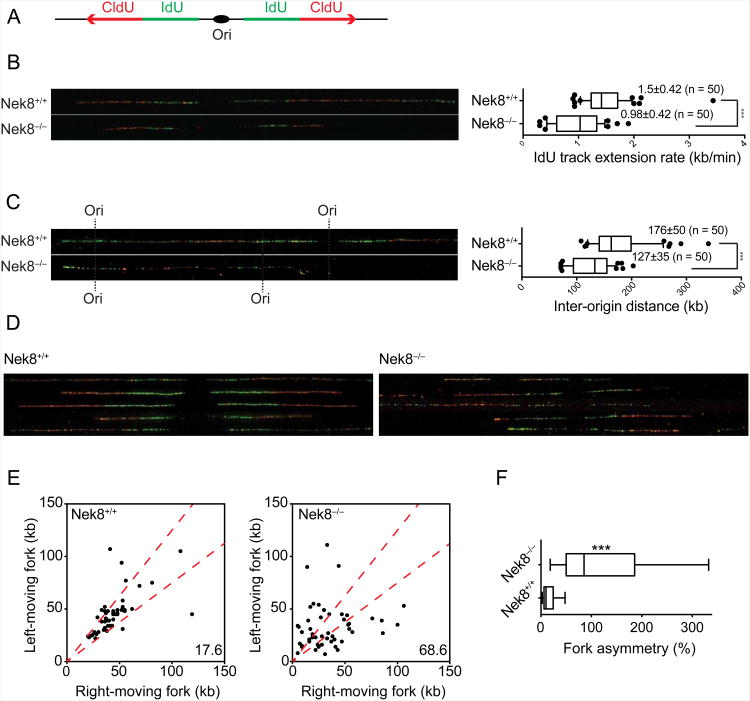
Figure 3.
NEK8 regulates origin firing, replication fork speed, and fork stability.
(A) Schematic of DNA combing. Primary MEFs were sequentially labeled with IdU (green) and CldU (red) for 30 min each. Individual DNA fibers were stretched and then visualized by immunofluorescence. Representative images of DNA fibers are presented. (B) Graph depicts the distribution of IdU track lengths (right panel). (C) Inter-origin distances were measured as the distance between two adjacent initiation sites during IdU pulse and median values are indicated in kb (right panel). (D) Representative images of DNA fibers show the fork asymmetry in Nek8−/− MEFs. (E) Scatter plot shows the length of left- and right-moving sister forks during CldU pulse. The central area marked with red lines indicates sister forks with less than a 25% length difference and the percentage of outliers is shown. (F) Fork asymmetry between left and right forks was scored as the ratio of the longest distance to the shortest covered during CldU labeling for each sister replication fork. Approximately 50 fibers were analyzed in each condition. Box and whiskers in all graphs indicate 25-75 and 10-90 percentiles, respectively. The lines represent the median values. Asterisks indicate the P-value of the statistical test (Mann-Whitney rank sum t-test, ***p < 0.0001). All the data are from three independent experiments. See also Figure S3.