Abstract
In higher eukaryotes, RAD51 functions as an essential protein in homologous recombination and recombinational repair of DNA double strand breaks. During these processes, RAD51 catalyzes homologous pairing between single-stranded DNA and double-stranded DNA. Japonica cultivars of rice (Oryza sativa) encode two RAD51 proteins, RAD51A1 and RAD51A2, whereas only one RAD51 exists in yeast and mammals. However, the functional differences between RAD51A1 and RAD51A2 have not been elucidated, because their biochemical properties have not been characterized. In the present study, we purified RAD51A1 and RAD51A2, and found that RAD51A2 robustly promotes homologous pairing in vitro. RAD51A1 also possesses homologous-pairing activity, but it is only about 10% of the RAD51A2 activity. Both RAD51A1 and RAD51A2 bind to ssDNA and dsDNA, and their DNA binding strictly requires ATP, which modulates the polymer formation activities of RAD51A1 and RAD51A2. These findings suggest that although both RAD51A1 and RAD51A2 have the potential to catalyze homologous pairing, RAD51A2 may be the major recombinase in rice.
Introduction
DNA double strand breaks (DSBs) are generated by DNA damaging agents, ionizing radiation, and collapsed DNA replication forks [1], [2]. Unrepaired DSBs cause chromosomal aberrations, leading to tumorigenesis or cell death. Homologous recombination is the error-free repair pathway for DSBs in mitosis [3], [4], [5], and is also required for the proper segregation of homologous chromosomes during the first meiotic cell division [6], [7].
In eukaryotes, RAD51, which is the central recombinase in homologous recombination, has been identified as a homologue of Escherichia coli RecA [8], [9], [10], [11]. DMC1, another RecA homologue, has also been found in eukaryotes [12], [13]. RAD51 is involved in both mitotic and meiotic homologous recombination [8], whereas DMC1 functions exclusively in meiosis [12]. During homologous recombination, RAD51 catalyzes homologous pairing, by which single-stranded DNA (ssDNA) forms new Watson-Crick base pairs (heteroduplex) with a complementary strand of homologous double-stranded DNA (dsDNA) in an ATP-dependent manner [14], [15], [16], [17], [18]. The meiosis-specific DMC1 also possesses homologous-pairing activity [19], [20], [21], [22]. In the homologous-pairing reaction, RAD51 and DMC1 first bind to ssDNA, and form helical nucleoprotein filaments. The resulting nucleoprotein filaments then bind to dsDNA, and facilitate heteroduplex formation in the ternary complex containing ssDNA, dsDNA, and either RAD51 or DMC1.
In Saccharomyces cerevisiae, strains bearing mutations in the Rad51 gene are quite sensitive to DNA damage agents, but are viable [3], [8]. In mice, the RAD51 gene knockout leads to early embryonic lethality [23], [24]. Similarly, deletion of the RAD51 gene in chicken DT40 cells causes the accumulation of chromosomal breaks and subsequent cell death [25]. These findings indicated that RAD51 is essential in higher eukaryotes. RAD51 has also been found in higher plants [26], [27], [28], [29] and a moss [30]. In maize, RAD51 facilitates proper homologous chromosome pairing in meiosis [31], [32]. The Arabidopsis genome has a single RAD51 gene [27], whereas the Zea mays and Physcomitrella patens genomes contain two genes [28], [30]. In Arabidopsis, the loss of RAD51 function has no impact on both the vegetative and flower development, and no abnormalities were detected in mitosis, whereas the mutant results in meiotic defects and causes sterility [33]. In contrast to Arabidopsis, the double knockout of two RAD51 genes significantly affects both the vegetative and generative development in Physcomitrella [34]. Since the two Physcomitrella RAD51 proteins reportedly promote homologous pairing with similar efficiencies [35], they may redundantly function in the moss.
In cultivated rice (Oryza sativa), a single copy of the RAD51 gene has been reported so far in an indica cultivar [36], while the japonica cultivar group genome contains two non-allelic RAD51A1 and RAD51A2 genes. Interestingly, the O. sativa genome also contains two DMC1 genes, DMC1A and DMC1B. Purified DMC1A and DMC1B promoted significant homologous recombination reactions in vitro [37], suggesting that both DMC1A and DMC1B are bona fide recombinases functioning in rice meiosis. However, the recombination activities of RAD51A1 and RAD51A2 from the japonica cultivar group have not been compared so far. In the present study, we purified the japonica rice RAD51A1 and RAD51A2 proteins, and found that RAD51A2 possesses robust homologous-pairing activity, which is about 10-fold higher than that of RAD51A1.
Materials and Methods
Protein Purification
The DNA fragments encoding the RAD51A1 and RAD51A2 sequences from the japonica cultivar Nipponbare (NCBI accession nos. AB080262 and AB080264, respectively) were cloned into the NdeI-BamHI sites of the pET-15b expression vector (Novagen). In each construct, the His6 tag-coding sequence was fused at the N-terminal end of the RAD51A1- and RAD51A2-coding sequences. His6-tagged RAD51A1 and RAD51A2 were individually expressed in the Escherichia coli BLR(DE3) pLysS strain, in which the recA gene has been deleted (Novagen). The cells producing the proteins were resuspended in buffer A [50 mM Tris-HCl (pH 8.0), 2 M NaCl, 10 mM EDTA, 5 mM 2-mercaptoethanol, and 10% glycerol], and were disrupted by sonication. The cell debris was removed by centrifugation for 20 min at 27,700 × g, and the supernatant was mixed gently with 3 ml of cOmplete His-Tag Purification Resin (Roche) at 4°C for 1 hr. The protein-bound beads were then packed into an Econo-column (Bio-Rad Laboratories), and were washed with 150 ml of buffer A, containing 20 mM imidazole. The proteins were eluted by a 60 ml linear gradient of imidazole from 20 to 300 mM. The peak fractions were collected, and 8 units of thrombin protease (GE Healthcare Biosciences) per mg of protein were added to remove the His6 tag. The samples were immediately dialyzed against a buffer containing 20 mM Tris-HCl (pH 8.0), 500 mM NaCl, 5 mM EDTA, 5 mM 2-mercaptoethanol, and 10% glycerol. After removal of the His6 tag, RAD51A1 and RAD51A2 were loaded onto a Superdex 200 gel filtration column (HiLoad 26/60 preparation grade; GE Healthcare Biosciences), which was previously equilibrated with buffer containing 20 mM Tris-HCl (pH 8.0), 2 M NaCl, 10 mM EDTA, 5 mM 2-mercaptoethanol, and 10% glycerol. RAD51A1 and RAD51A2 were eluted from the Superdex 200 gel filtration column, dialyzed against buffer containing 20 mM HEPES-NaOH (pH 7.5), 400 mM NaCl, 0.1 mM EDTA, 2 mM 2-mercaptoethanol, and 10% glycerol, and stored at −80°C. The DNA fragments encoding the RAD51A1(A2L2) and RAD51A2(A1L2) mutants were constructed, and were cloned into the NdeI-BamHI sites of the pET-15b expression vector (Novagen). The RAD51A1(A2L2) and RAD51A2(A1L2) mutants were then purified by the same method as for the wild type proteins. Human RAD51 was expressed in E. coli cells [38], and was purified as described previously [39].
DNA Substrates
Single-stranded φX174 viral (+) strand DNA and double-stranded φX174 replicative form I DNA were purchased from New England Biolabs, and the linear dsDNA was prepared from the φX174 replicative form I DNA by PstI digestion. In the D-loop formation assay, superhelical dsDNAs were prepared by a method avoiding alkaline treatment of the cells harboring the plasmid DNA [40]. For the ssDNA substrate used in the D-loop formation assay, the following HPLC-purified DNA oligonucleotides were purchased from Nihon Gene Research Laboratory: 50-mer, 5′-ATT TCA TGC TAG ACA GAA GAA TTC TCA GTA ACT TCT TTG TGC TGT GTG TA-3′. All of the DNA concentrations are expressed in moles of nucleotides.
The ATPase Assay
The ATPase activities of the proteins were analyzed by the release of 32Pi from [γ-32P]ATP. Rice RAD51A1 or RAD51A2 (1.5 µM) was incubated at 37°C in the presence of φX174 circular ssDNA (20 µM) or linearized φX174 dsDNA (20 µM), or in the absence of DNA, in 10 µl of reaction buffer, containing 24 mM HEPES-NaOH (pH 7.5), 80 mM NaCl, 0.02 mM EDTA, 0.4 mM 2-mercaptoethanol, 2% glycerol, 1 mM MgCl2, 1 mM DTT, 5 or 500 µM ATP, 5 nCi [γ-32P]ATP, and 0.1 mg/ml BSA. At the indicated times, the reaction was stopped by the addition of 1 µl of 0.5 M EDTA, and the products were separated by thin layer chromatography on polyethyleneimine-cellulose in a 0.5 M LiCl and 1 M formic acid solution.
The D-loop Formation Assay
The indicated amount of rice RAD51A1, RAD51A2, or human RAD51 was incubated with the 32P-labeled 50-mer oligonucleotide (1 µM) in the presence or absence of CaCl2 (1 mM) at 37°C for 5 min, in 9 µl of reaction buffer, containing 24 mM HEPES-NaOH (pH 7.5), 80 mM NaCl, 0.02 mM EDTA, 0.4 mM 2-mercaptoethanol, 2% glycerol, 1 mM MgCl2, 1 mM DTT, 1 mM ATP, 0.1 mg/ml BSA, 20 mM creatine phosphate, and 75 µg/ml creatine kinase. The reactions were then initiated by the addition of 1 µl of pGsat4 superhelical dsDNA (30 µM), and were continued at 37°C for 5 min. The reactions were stopped by the addition of 0.2% SDS and 1.5 mg/ml proteinase K (Roche), and were further incubated at 37°C for 15 min. After adding 6-fold loading dye, the deproteinized reaction products were separated by 1% agarose gel electrophoresis in 1× TAE buffer (40 mM Tris-acetate (pH 8.0), and 1 mM EDTA) at 4 V/cm for 2 hrs. The gels were dried and exposed to an imaging plate. The gel images were visualized using an FLA−7000 imaging analyzer (Fujifilm). We confirmed that both RAD51A1 and RAD51A2 did not promote the D-loop formation with a heterologous dsDNA (pB5Sarray, which contained 11 repeats of a sea urchin 5S rRNA gene (207-bp fragment) within the pBlueScript II SK(+) vector) under the conditions employed in the present study.
The DNA-binding Assay
The φX174 circular ssDNA (20 µM) or the linearized φX174 dsDNA (20 µM) was mixed with rice RAD51A1, RAD51A2, or human RAD51 in the presence or absence of 1 mM ATP, in 10 µl of a standard reaction solution, containing 30 mM HEPES-NaOH (pH 7.5), 200 mM NaCl, 0.05 mM EDTA, 1 mM 2-mercaptoethanol, 5% glycerol, 1 mM MgCl2, 1 mM DTT, and 0.1 mg/ml BSA. The reaction mixtures were incubated at 37°C for 10 min, and were then separated by 0.8% agarose gel electrophoresis in 1× TAE buffer at 3.3 V/cm for 2 hrs. The bands were visualized by ethidium bromide staining.
Electron Microscopy
Rice RAD51A1 or RAD51A2 (0.5 µM) was mixed with the φX174 dsDNA (0.5 µM) in 20 µl of reaction buffer, containing 30 mM HEPES-NaOH (pH 7.5), 200 mM NaCl, 0.05 mM EDTA, 1 mM 2-mercaptoethanol, 5% glycerol, 1 mM MgCl2, and 1 mM ATP, and was incubated at 37°C for 10 min. Samples (3 µl) were adsorbed on a carbon grid and stained with 2% uranium acetate. The samples were examined with a JEM2000FX electron microscope (JOEL).
Gel Filtration Analysis
Rice RAD51A1 or RAD51A2 (70 µg) was incubated with or without ATP (1 mM) or ADP (1 mM) at room temperature for 30 min, in a reaction mixture containing 20 mM HEPES-NaOH (pH 7.5), 355 mM NaCl, 0.1 mM EDTA, 2 mM 2-mercaptoethanol, 10% glycerol, and 1 mM MgCl2. The samples were then analyzed by Superdex 200 GL 10/300 (GE Healthcare BioSciences) gel filtration chromatography. The elution buffer contained 20 mM HEPES-NaOH (pH 7.5), 400 mM NaCl, 0.1 mM EDTA, 2 mM 2-mercaptoethanol, and 1 mM MgCl2, and the elution profiles of RAD51A1 or RAD51A2 were monitored by UV absorption at 280 nm.
The pull-down Assay with Ni-NTA Beads
Purified His6-tagged RAD51A1 (2.0 µg) was mixed with RAD51A1 (4.0 µg) or RAD51A2 (4.0 µg) in 60 µl of binding buffer, containing 19 mM HEPES-NaOH (pH 7.5), 380 mM NaCl, 0.095 mM EDTA, 1.9 mM 2-mercaptoethanol, 0.025% Triton X-100, and 9.5% glycerol, and then Ni-NTA agarose beads (1.5 µl, 50% slurry) were added. After an incubation at room temperature for 1 hr, the beads were washed two times with 300 µl of wash buffer, containing 20 mM HEPES-NaOH (pH 7.5), 400 mM NaCl, 0.1 mM EDTA, 2 mM 2-mercaptoethanol, and 10% glycerol. The proteins bound to the beads were fractionated by 10% SDS-PAGE, and the bands were visualized by Coomassie Brilliant Blue staining.
Results
Purification of Rice RAD51A1 and RAD51A2
Two RAD51 genes, RAD51A1 and RAD51A2, have been identified in rice (japonica cultivar group). Only a single RAD51 gene has been reported in an indica cultivar (Pusa Basmati 1) [36]. The amino acid sequence of the indica RAD51 is more similar to the japonica RAD51A2 (98%) than to RAD51A1 (93%) (Figure 1A).
Figure 1. Purification of rice RAD51A1 and RAD51A2.
(A) The amino acid sequences of rice RAD51A1 and RAD51A2 from japonica cultivar group, cv. Nipponbare, rice RAD51 from indica cultivar group, cv. Pusa Basmati 1, and human RAD51, aligned with the ClustalX software [50]. Black and gray boxes indicate identical and similar amino acid residues, respectively. The L1 and L2 loops, which are important for DNA binding, are represented by red lines. (B) Purified rice RAD51A1, RAD51A2, and human RAD51. Lane 1 indicates the molecular mass markers, and lanes 2, 3, and 4 represent rice RAD51A1 (0.5 µg), RAD51A2 (0.5 µg), and human RAD51 (0.5 µg), respectively. (C) The ATPase activities of Oryza sativa RAD51A1 and RAD51A2. The reactions were conducted with φX174 circular ssDNA (left panel), linearized φX174 dsDNA (center panel), or without DNA (right panel), in the presence of 5 µM ATP. Blue circles and red squares represent the experiments with RAD51A1 and RAD51A2, respectively. The averages of three independent experiments are shown with the SD values.
RAD51A1 and RAD51A2 share 92% amino acid identity with each other, and both exhibit 69% amino acid identity with human RAD51, respectively (Figure 1A). To reveal the functional differences between RAD51A1 and RAD51A2, we expressed japonica rice RAD51A1 and RAD51A2 as recombinant proteins in Escherichia coli cells, and purified them by a three-step purification method (Figure 1B, lanes 2 and 3). We used E. coli cells lacking the recA gene, to eliminate the possibility of contamination with the RecA protein. The His6 tag was removed from the RAD51A1 and RAD51A2 portions by thrombin protease treatment, during the purification procedure.
The purified RAD51A1 and RAD51A2 hydrolyzed ATP in ssDNA- and dsDNA-dependent manners (Figure 1C). RAD51A2 exhibited higher ATP hydrolyzing activity than RAD51A1 in the presence of ssDNA or dsDNA (Figure 1C). The ATP hydrolyzing activities of RAD51A1 and RAD51A2 were lower than that of human RAD51 (Figure 1C). Under the experimental conditions used in this study, the kcat values of the ssDNA-dependent ATP hydrolyzing activities of RAD51A1, RAD51A2, and human RAD51 were about 0.06, 0.08, and 0.1 min−1, respectively.
Rice RAD51A2 Exhibits Robust Homologous-pairing Activity
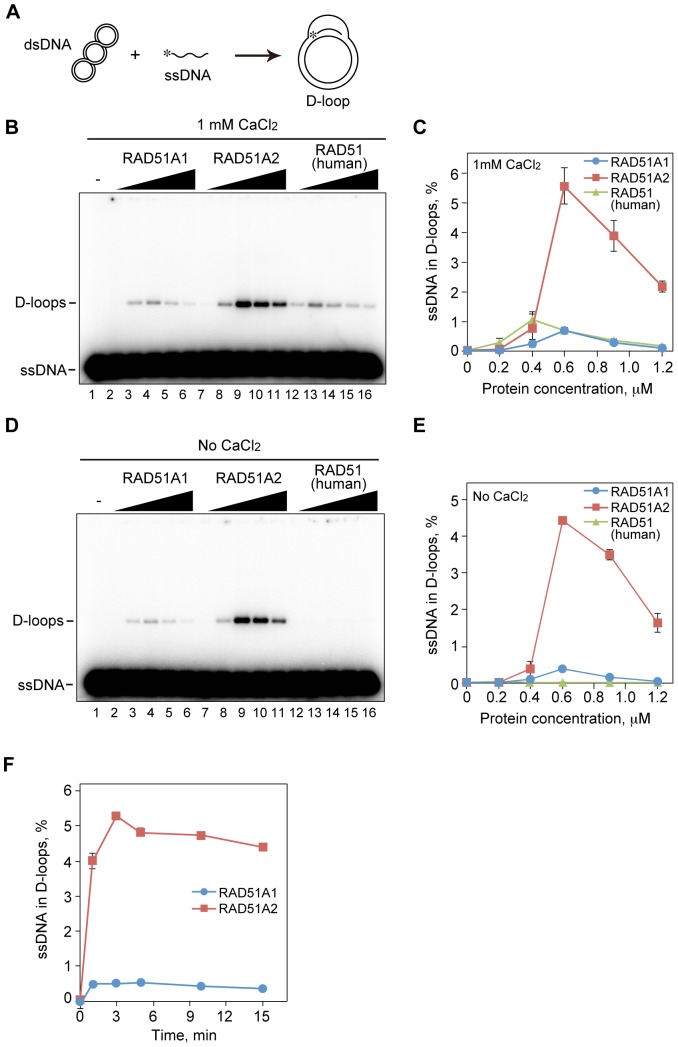
We next tested the homologous-pairing activities of RAD51A1 and RAD51A2. To this end, we performed D-loop formation assays. In these assays, D-loops were detected as a homologous pairing product, in which a 32P-labeled ssDNA fragment was hybridized to the complementary strand of superhelical dsDNA (Figure 2A). The homologous pairing mediated by the RecA-family proteins is an ATP- and Mg2+-dependent reaction [41], [42]. In addition, Ca2+ions reportedly stimulate the homologous pairing by human RAD51 [43]. Therefore, we conducted the homologous-pairing reactions in the presence of ATP, Mg2+, and Ca2+. We found that RAD51A2 robustly catalyzed homologous pairing, as compared to human RAD51 (Figure 2B and C). On the other hand, RAD51A1 exhibited the homologous-pairing activity, but it was only about 10% of the RAD51A2 activity under the conditions with ATP, Mg2+, and Ca2+(Figure 2B and C). Interestingly, RAD51A1 and RAD51A2 did not require Ca2+to catalyze homologous pairing, although the homologous-pairing activity of human RAD51 was hardly detected in the absence of Ca2+(Figure 2D and E). The homologous-pairing activity of human RAD51 was somewhat reduced under the 80 mM NaCl conditions employed for RAD51A1 and RAD51A2 (data not shown). RAD51A2 still exhibited about 12-fold higher homologous-pairing activity than RAD51A1 under the conditions without Ca2+ion (Figure 2D and E). In addition, time course experiments revealed that the D-loop formation by RAD51A2 became rapidly saturated within 3 min, and RAD51A2 possesses higher homologous-pairing activity than RAD51A1 (Figure 2F).
Figure 2. The homologous pairing activities of rice RAD51A1 and RAD51A2.
(A) A schematic representation of the D-loop formation assay. Asterisks indicate the 32P-labeled end of the 50-mer ssDNA. (B) The D-loop formation assay in the presence of Ca2+(Protein titration experiments). The indicated amounts of rice RAD51A1, RAD51A2, or human RAD51 were incubated with the 32P-labeled 50-mer ssDNA, and the homologous-pairing reaction was initiated by the addition of superhelical dsDNA. Reactions were allowed to proceed for 5 min. Lane 1 indicates a negative control experiment without protein, and lanes 2–6, 7–11, and 12–16 represent the reactions conducted with RAD51A1, RAD51A2, and human RAD51, respectively. The protein concentrations were 0.2 µM (lanes 2, 7, and 12), 0.4 µM (lanes 3, 8, and 13), 0.6 µM (lanes 4, 9, and 14), 0.9 µM (lanes 5, 10, and 15), and 1.2 µM (lanes 6, 11, and 16). (C) Graphic representation of the experiments shown in panel B. The averages of three independent experiments are shown with the SD values. Blue circles, red squares, and green triangles represent the experiments with RAD51A1, RAD51A2, and human RAD51, respectively. (D) The D-loop formation assay without Ca2+(Protein titration experiments). The homologous pairing reactions were conducted without Ca2+, and were performed according to the same procedure as shown in panel B. (E) Graphic representation of the experiments shown in panel D. The averages of three independent experiments are shown with the SD values. Blue circles, red squares, and green triangles represent the experiments with RAD51A1, RAD51A2, and human RAD51, respectively. (F) Graphic representation of the D-loop formation assay (time course experiments). The homologous pairing reactions were conducted according to the same procedure as shown in panel B. Rice RAD51A1 (0.5 µM) or RAD51A2 (0.5 µM) was used in the time course experiments.
The DNA-binding Activities of Rice RAD51A1 and RAD51A2
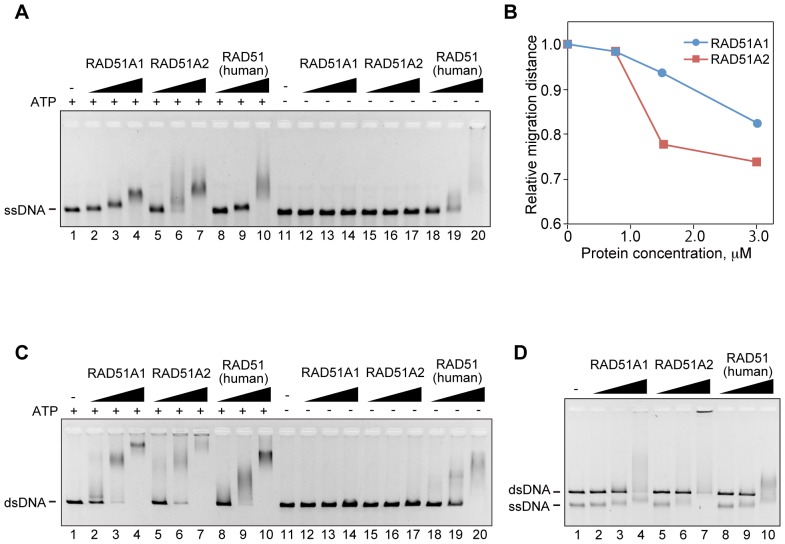
We next tested the DNA-binding activities of RAD51A1 and RAD51A2. As shown in Figure 3A and C, both RAD51A1 and RAD51A2 bound to ssDNA and dsDNA, as well as human RAD51, in the presence of ATP (lanes 1–10). Interestingly, we found that RAD51A1 and RAD51A2 strictly required ATP for their ssDNA- and dsDNA-binding activities (Figure 3A and C, lanes 11–17), unlike human RAD51 (Figure 3A and C, lanes 18–20). Consistently, RAD51A1 and RAD51A2 formed helical nucleoprotein filaments with dsDNA in the presence of ATP (Figure 4A and B), but the filaments were not detected in the absence of ATP, due to their inability to bind DNA (Figure 3A and C). The average helical pitches of both the RAD51A1 and RAD51A2 filaments were about 9.15 nm. These results suggested that ATP modifies the polymerization states of RAD51A1 and RAD51A2, and enhances their DNA-binding activities.
Figure 3. The DNA-binding activities of rice RAD51A1 and RAD51A2.
Circular φX174 ssDNA (20 µM) (A) or linear φX174 dsDNA (20 µM) (C) was incubated with rice RAD51A1, RAD51A2, or human RAD51 at 37°C for 10 min. The samples were then separated by 0.8% agarose gel electrophoresis in TAE buffer, and were visualized by ethidium bromide staining. Lanes 1–10 and 11–20 represent the reactions conducted with and without ATP, respectively. Lanes 1 and 11 indicate negative control experiments without protein. Lanes 2–4 and 12–14 represent the experiments conducted with RAD51A1. Lanes 5–7 and 15–17 represent the experiments conducted with RAD51A2. Lanes 8–10 and 18–20 represent the experiments conducted with human RAD51. The protein concentrations were 0.75 µM (lanes 2, 5, 8, 12, 15, and 18), 1.5 µM (lanes 3, 6, 9, 13, 16, and 19) and 3 µM (lanes 4, 7, 10, 14, 17, and 20). (B) Graphic representation of the relative migration distances of the RAD51A1- and RAD51A2-ssDNA complexes. The migration distances relative to the free DNA are plotted against the protein concentrations. (D) Competitive ssDNA- and dsDNA-binding. Circular φX174 ssDNA (20 µM) and linear φX174 dsDNA (20 µM) were incubated with rice RAD51A1, RAD51A2, or human RAD51 at 37°C for 10 min, under the 120 mM NaCl conditions. The samples were then separated by 0.8% agarose gel electrophoresis in TAE buffer, and were visualized by ethidium bromide staining. Lane 1 indicates negative control experiments without protein. Lanes 2–4, 5–7, and 8–10 represent the experiments conducted with RAD51A1, RAD51A2, and human RAD51, respectively. The protein concentrations were 0.9 µM (lanes 2, 5, and 8), 1.8 µM (lanes 3, 6, and 9), and 3.6 µM (lanes 4, 7, and 10).
Figure 4. Electron microscopic images of RAD51A1 and RAD51A2 complexed with DNA.

(A and B) Electron microscopic images of rice RAD51A1 (A) and RAD51A2 (B) filaments formed on the φX174 dsDNA in the presence of ATP. The average helical pitches of the RAD51A1 and RAD51A2 filaments were about 9.15 nm. The black bar denotes 100 nm.
Interestingly, the RAD51A2-ssDNA complexes migrated more slowly than the RAD51A1-ssDNA complexes (Figure 3A and B), suggesting that RAD51A2 may have higher ssDNA-binding activity than RAD51A1. In addition, competitive DNA binding assays revealed that RAD51A2, but not RAD51A1, significantly preferred to bind ssDNA (Figure 3D). The higher ssDNA-binding activity of RAD51A2 may be a major reason for its higher homologous-pairing activity (Figure 2).
ATP-dependent Polymer Formation by Rice RAD51A1 and RAD51A2
To test whether ATP affects the polymerization states of RAD51A1 and RAD51A2, we tested the polymerization activities of RAD51A1 and RAD51A2 with or without ATP. To do so, we performed gel filtration chromatography. As shown in Figure 5 (top panels), RAD51A1 and RAD51A2 eluted at the apparent molecular sizes corresponding to 8-mer and 5-mer, respectively, without ATP. As expected, in the presence of ATP, the apparent molecular sizes of RAD51A1 and RAD51A2 drastically increased (Figure 5A and B, middle panels). These ATP-dependent changes in the RAD51A1 and RAD51A2 polymer formation were not induced by ADP (Figure 5A and B, bottom panels). Therefore, ATP actually enhances the oligomerization activities of RAD51A1 and RAD51A2. These ATP-dependent changes in the polymers formed by RAD51A1 and RAD51A2 may stimulate their DNA-binding activities during homologous pairing.
Figure 5. The polymerization activities of rice RAD51A1 and RAD51A2.
A 70 µg portion of RAD51A1 (A) or RAD51A2 (B) was analyzed by Superdex 200 gel filtration chromatography. The top, middle, and bottom panels represent the experiments without ATP, with ATP, and with ADP, respectively.
Rice RAD51A1 and RAD51A2 may not form a co-filament
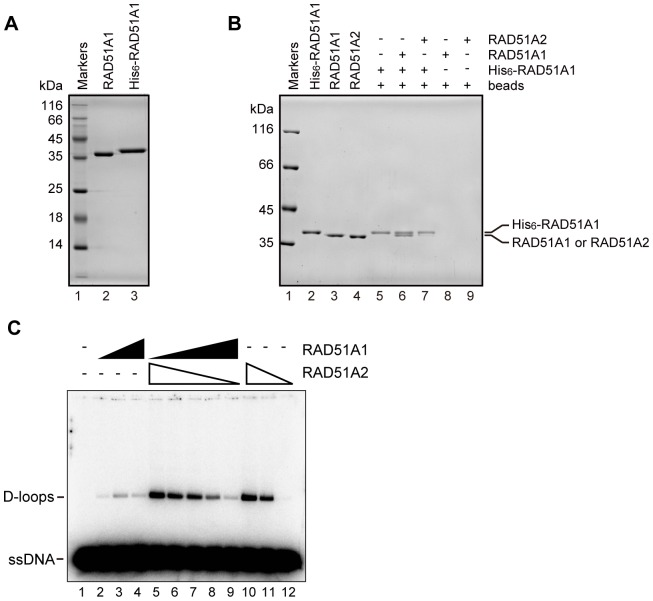
To test whether RAD51A1 and RAD51A2 form a co-filament, we performed the Ni-bead pull-down assay. In this assay, the His6-tagged RAD51A1 was purified (Figure 6A), and incubated with RAD51A1 or RAD51A2. The RAD51A1 or RAD51A2 bound to the His6-tagged RAD51A1 was detected. As shown in Figure 6B, RAD51A1 was efficiently pulled down by His6-tagged RAD51A1, but only a trace amount of RAD51A2 was detected. These results suggested that RAD51A1 efficiently binds to RAD51A1, but not to RAD51A2. Consistently, the homologous-pairing activities of RAD51A1 and RAD51A2 were not synergistically affected in the presence of various amounts of RAD51A1 and RAD51A2 (Figure 6C). Therefore, we concluded that rice RAD51A1 and RAD51A2 may not form a co-filament, and thus distinctly function in rice.
Figure 6. Interactions of rice RAD51A1 and RAD51A2.
(A) Purified His6-tagged RAD51A1 protein. Lane 1 indicates the molecular mass markers, and lanes 2 and 3 represent the RAD51A1 and His6-tagged RAD51A1 proteins (0.5 µg). (B) The pull-down assay with Ni–NTA beads. Lane 1 represents molecular mass markers. Lanes 2, 3, and 4 show purified protein controls of His6-tagged RAD51A1, RAD51A1, and RAD51A2, respectively. Lane 5 indicates a negative control experiment without RAD51A1 and RAD51A2, in the presence of His6-tagged RAD51A1. Lanes 8 and 9 indicate negative control experiments with RAD51A1 and RAD51A2, respectively, in the absence of His6-tagged RAD51A1. Lanes 6 and 7 indicate experiments with RAD51A1 and RAD51A2, respectively, in the presence of His6-tagged RAD51A1. The proteins bound to His6-tagged RAD51A1 were pulled down by the Ni–NTA agarose beads. The samples were fractionated by 10% SDS–PAGE, and the protein bands were visualized by Coomassie Brilliant Blue staining. (C) The D-loop formation assay in the presence of Ca2+(Protein titration experiments). The indicated amounts of rice RAD51A1 and RAD51A2 were mixed and incubated with the 32P-labeled 50-mer ssDNA, and the homologous-pairing reaction was initiated by the addition of superhelical dsDNA. Reactions were allowed to proceed for 5 min. Lane 1 indicates a negative control experiment without protein, and lanes 2–4 and 10–12 indicate positive control experiments with RAD51A1 and RAD51A2, respectively. The protein concentrations were 0.2 µM (lanes 2 and 12), 0.4 µM (lanes 3 and 11), and 0.6 µM (lanes 4 and 10). Lanes 5–9 represent experiments with various amounts of RAD51A1 and RAD51A2. Lane 5: RAD51A1 (0 µM) and RAD51A2 (0.6 µM). Lane 6: RAD51A1 (0.2 µM) and RAD51A2 (0.4 µM). Lane 7: RAD51A1 (0.3 µM) and RAD51A2 (0.3 µM). Lane 8: RAD51A1 (0.4 µM) and RAD51A2 (0.2 µM). Lane 9: RAD51A1 (0.6 µM) and RAD51A2 (0 µM).
Contribution of the L2 Loop Regions of Rice RAD51A1 and RAD51A2 to the Homologous-pairing Activity
Two flexible loops, L1 and L2, of human RAD51 are known to be important for its DNA-binding and homologous-pairing activities [44]. There are no differences in the amino acid sequences of the L1 loop regions between RAD51A1 and RAD51A2 (Figure 1A). In contrast, in the L2 loop region, the Ser276 residue of RAD51A1 is substituted with Ala279 in RAD51A2 (Figure 1A). In addition, one amino acid insertion, at the Ala280 residue of RAD51A1, is found in the L2 loop region (Figure 1A). These amino acid variations in the L2 loops region may be responsible for the differences in homologous pairing between RAD51A1 and RAD51A2.
Therefore, we constructed and purified the RAD51A1(A2L2) and RAD51A2(A1L2) mutants (Figure 7A), in which the L2 regions of RAD51A1 and RAD51A2 were replaced by the RAD51A2-L2 and RAD51A1-L2 regions, respectively. We then tested their homologous-pairing activities. Interestingly, the homologous-pairing activity of RAD51A1(A2L2) was clearly higher than that of the wild type RAD51A1 (Figure 7B, lanes 10–13). On the other hand, RAD51A2(A1L2) exhibited reduced homologous-pairing activity, as compared to the wild type RAD51A2 (Figure 7B, lanes 14–17). These results indicated that the L2 regions of RAD51A1 and RAD51A2 are at least partly responsible for their respective homologous-pairing activities.
Figure 7. The homologous-pairing activities of the RAD51A1(A2L2) and RAD51A2(A1L2) mutants.
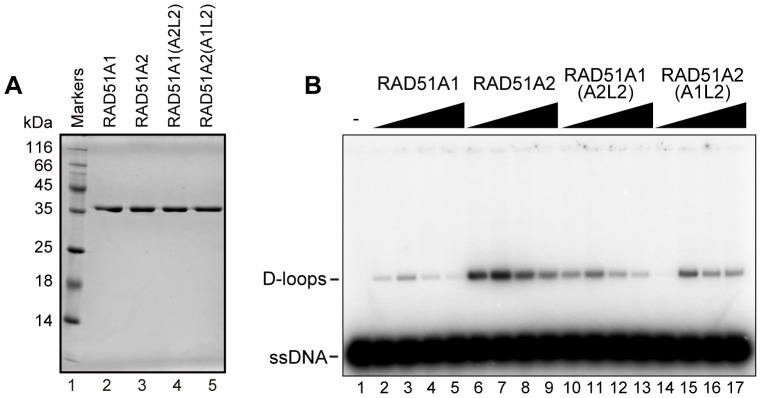
(A) Purified RAD51A1(A2L2) and RAD51A2(A1L2) mutants. Lane 1 indicates the molecular mass markers, and lanes 2 and 3 represent RAD51A1 and RAD51A2, respectively. Lanes 4 and 5 represent RAD51A1(A2L2) and RAD51A2(A1L2). An aliquot (0.5 µg) of each protein was analyzed. (B) The D-loop formation assay (Protein titration experiments). The indicated amounts of rice RAD51A1, RAD51A2, RAD51A1(A2L2), or RAD51A2(A1L2) were incubated with the 32P-labeled 50-mer ssDNA, and the homologous-pairing reaction was initiated by the addition of superhelical dsDNA. Reactions were allowed to proceed for 5 min. Lane 1 indicates a negative control experiment without protein, and lanes 2–5, 6–9, 10–13, and 14–17 represent the reactions conducted with RAD51A1, RAD51A2, RAD51A1(A2L2), and RAD51A2(A1L2), respectively. The protein concentrations were 0.4 µM (lanes 2, 6, 10, and 14), 0.6 µM (lanes 3, 7, 11, and 15), 0.9 µM (lanes 4, 8, 12, and 16), and 1.2 µM (lanes 5, 9, 13, and 17).
Discussion
Two RAD51 proteins, RAD51A1 and RAD51A2, have been identified in japonica cultivars of rice. A previous study revealed that the amount of RAD51A2 mRNA, but not that of RAD51A1 mRNA, is increased by ionizing irradiation, which induces DSBs [45]. The mRNA level of RAD51A2 is higher than that of RAD51A1 in embryogenic calli of rice, which exhibit higher recombination rates, as compared to other organs [46]. These facts suggested that RAD51A2 may be the major functional recombinase in the RAD51-mediated DSB repair and homologous recombination in rice. A RAD51 gene has been identified in the rice indica-cultivar group [36]. The indica RAD51 shares 98% amino acid identity with the japonica RAD51A2, and 93% identity with the japonica RAD51A1. Since an additional locus for RAD51, which displays higher homology to RAD51A1 than RAD51A2, has been found in the genome sequence of the indica rice cultivar Kasalath (J. Wu, personal communication), it is quite likely that two genes for RAD51 also exist in the indica cultivar group. We determined that RAD51A2 possesses robust homologous-pairing activity (Figure 2). RAD51A1 also catalyzes homologous pairing, but its activity is quite low, as compared to RAD51A2 (Figure 2). These biochemical findings are consistent with the idea that RAD51A2 is the major functional recombinase in rice.
However, the homologous-pairing activity of RAD51A1 may not be negligible, because it is comparable to that of human RAD51 (Figure 2). Therefore, RAD51A1 may not simply be a non-functional paralogue. Although the functional reasons for the diversity of RAD51A1 and RAD51A2 in rice have not been clarified yet, the biochemical distinctions and similarities of these two rice recombinases revealed in the present study may provide important insights into understanding their biological significance.
We found that RAD51A1 and RAD51A2 bind to ssDNA and dsDNA in ATP-dependent manners. These findings suggested that ATP may stabilize the protein-protein interactions within the RAD51A1 and RAD51A2 filaments. The crystal structures of the RadA protein, an archaeal RAD51 orthologue, and the RecA-DNA complexes revealed that the ATP molecule binds to the protein-protein interfaces within the filaments [47], [48]. The ATP bound to RAD51A1 and RAD51A2 may convert the RAD51 structures, enhancing the interactions between RAD51 monomers within the filament. Consistent with this idea, we found that ATP drastically changes the polymerization states of RAD51A1 and RAD51A2 (Figure 5). A similar ATP-dependent change in polymerization was also observed for human RAD51 (data not shown). In addition, the electrophoretic gel mobilities of the human RAD51-DNA complexes differ in the presence and absence of ATP, although human RAD51 binds DNA without ATP (Figure 3). The function of ATP in the formation of functional RAD51 filaments has been clarified with the rice RAD51A1 and RAD51A2 proteins.
In the present study, we detected significant differences in homologous pairing between RAD51A1 and RAD51A2, although they share high amino acid identity (92%). These biochemical differences may be a consequence of the amino acid differences in the DNA-binding regions. Bacterial RecA contains two flexible loops, L1 and L2, which are involved in DNA binding, and the corresponding L1 and L2 loops of human RAD51 are also important for its DNA-binding and recombination activities [44]. Although the amino acid sequences are the same in the L1 loop regions of RAD51A1 and RAD51A2, one amino acid difference (Ser276 in RAD51A1 and Ala279 in RAD51A2) and one amino acid insertion (Ala280) in RAD51A1 are found in the L2 loop region. In addition, it has been reported that the N-terminal fragment of human RAD51 (amino acids 1–114) directly binds to DNA [49]. Many amino acid differences exist in the corresponding N-terminal regions of RAD51A1 and RAD51A2. Therefore, these amino acid differences in the L2 loops and N-terminal regions may be responsible for the differences in homologous pairing between RAD51A1 and RAD51A2. In the present study, the D-loop formation assay with the L2-swapping mutants, RAD51A1(A2L2) and RAD51A2(A1L2), revealed that the L2 regions are actually responsible for their respective homologous-pairing activities. Intriguingly, the L2 loop sequence of japonica RAD51A2 is perfectly conserved with that of indica RAD51, which reportedly promotes homologous pairing [36], although the N-terminal sequence of indica RAD51 differs from those of both japonica RAD51A1 and RAD51A2. In fact, the L2-swapping mutations did not restore the homologous-pairing activities of wild type RAD51A1 and RAD51A2, indicating that the regions outside the L2 loop may also be important for the distinctive RAD51A1 and RAD51A2 activities. In addition, several amino acid differences also exist between RAD51A1 and RAD51A2 outside these DNA-binding regions. In a future study, it will be quite interesting to identify the amino acid residues responsible for the functional differences between RAD51A1 and RAD51A2.
Acknowledgments
We are grateful to Dr. Jianzhong Wu, at the National Institute of Agrobiological Sciences in Tsukuba, Japan, for communicating unpublished results. We also thank S. Machida (Waseda University) and W. Kobayashi (Waseda University) for discussions and manuscript preparation. HK is a research fellow in the Waseda Research Institute for Science and Engineering.
Funding Statement
This work was partly supported by grants from the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) and from the Bio-Oriented Technology Research Advancement Institution (BRAIN) in Tokyo to TS and HK, and to HI and ST, respectively. This work was also supported in part by Grants-in-Aid from the Japanese Society for the Promotion of Science (JSPS) and the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Whitaker SJ (1992) DNA damage by drugs and radiation: what is important and how is it measured? Eur J Cancer 28: 273–276. [DOI] [PubMed] [Google Scholar]
- 2. Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, et al. (2000) The importance of repairing stalled replication forks. Nature 404: 37–41. [DOI] [PubMed] [Google Scholar]
- 3. Symington LS (2002) Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev 66: 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. West SC (2003) Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol 4: 435–445. [DOI] [PubMed] [Google Scholar]
- 5. San Filippo J, Sung P, Klein H (2008) Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77: 229–257. [DOI] [PubMed] [Google Scholar]
- 6. Petronczki M, Siomos MF, Nasmyth K (2003) Un ménage à quatre: the molecular biology of chromosome segregation in meiosis. Cell 112: 423–440. [DOI] [PubMed] [Google Scholar]
- 7. Neale MJ, Keeney S (2006) Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature 442: 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shinohara A, Ogawa H, Ogawa T (1992) Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69: 457–470. [DOI] [PubMed] [Google Scholar]
- 9. Aboussekhra A, Chanet R, Adjiri A, Fabre F (1992) Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol Cell Biol 12: 3224–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Basile G, Aker M, Mortimer RK (1992) Nucleotide sequence and transcriptional regulation of the yeast recombinational repair gene RAD51 . Mol Cell Biol 12: 3235–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shinohara A, Ogawa H, Matsuda Y, Ushio N, Ikeo K, et al. (1993) Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA . Nat Genet 4: 239–243. [DOI] [PubMed] [Google Scholar]
- 12. Bishop DK, Park D, Xu L, Kleckner N (1992) DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69: 439–456. [DOI] [PubMed] [Google Scholar]
- 13. Habu T, Taki T, West A, Nishimune Y, Morita T (1996) The mouse and human homologs of DMC1, the yeast meiosis-specific homologous recombination gene, have a common unique form of exon-skipped transcript in meiosis. Nucleic Acids Res 24: 470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sung P (1994) Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265: 1241–1243. [DOI] [PubMed] [Google Scholar]
- 15. Sung P, Robberson DL (1995) DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell 82: 453–461. [DOI] [PubMed] [Google Scholar]
- 16. Baumann P, Benson FE, West SC (1996) Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell 87: 757–766. [DOI] [PubMed] [Google Scholar]
- 17. Maeshima K, Morimatsu K, Horii T (1996) Purification and characterization of XRad51.1 protein, Xenopus RAD51 homologue: recombinant XRad51.1 promotes strand exchange reaction. Genes Cells 1: 1057–1068. [DOI] [PubMed] [Google Scholar]
- 18. Gupta RC, Bazemore LR, Golub EI, Radding CM (1997) Activities of human recombination protein Rad51. Proc Natl Acad Sci USA 94: 463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Z, Golub EI, Gupta R, Radding CM (1997) Recombination activities of HsDmc1 protein, the meiotic human homolog of RecA protein. Proc Natl Acad Sci USA 94: 11221–11226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hong EL, Shinohara A, Bishop DK (2001) Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single-strand DNA (ssDNA) and assimilation of ssDNA into homologous super-coiled duplex DNA. J Biol Chem 276: 41906–41912. [DOI] [PubMed] [Google Scholar]
- 21. Kinebuchi T, Kagawa W, Enomoto R, Tanaka K, Miyagawa K, et al. (2004) Structural basis for octameric ring formation and DNA interaction of the human homologous-pairing protein Dmc1. Mol Cell 14: 363–374. [DOI] [PubMed] [Google Scholar]
- 22. Sehorn MG, Sigurdsson S, Bussen W, Unger VM, Sung P (2004) Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange. Nature 429: 433–437. [DOI] [PubMed] [Google Scholar]
- 23. Lim DS, Hasty P (1996) A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53 . Mol Cell Biol 16: 7133–7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, et al. (1996) Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci USA 93: 6236–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sonoda E, Sasaki MS, Buerstedde JM, Bezzubova O, Shinohara A, et al. (1998) Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J 17: 598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stassen NY, Logsdon JM, Vora GJ, Offenberg HH, Palmer JD, et al. (1997) Isolation and characterization of rad51 orthologs from Coprinus cinereus and Lycopersicon esculentum, and phylogenetic analysis of eukaryotic recA homologs. Curr Genet 31: 144–157. [DOI] [PubMed] [Google Scholar]
- 27. Doutriaux MP, Couteau F, Bergounioux C, White C (1998) Isolation and characterization of the RAD51 and DMC1 homologues from Arabidopsis thaliana . Mol Gen Genet 257: 283–291. [DOI] [PubMed] [Google Scholar]
- 28. Franklin AE, McElver J, Sunjevaric I, Rothstein R, Bowen B, et al. (1999) Three-dimensional microscopy of the Rad51 recombination protein during meiotic prophase. Plant Cell 11: 809–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Devisetty UK, Mayes K, Mayes S (2010) The RAD51 and DMC1 homoeologous genes of bread wheat: cloning, molecular characterization and expression analysis. BMC Res Notes 3: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Markmann-Mulisch U, Hadi MZ, Koepchen K, Alonso JC, Russo VEA, et al. (2002) The organization of Physcomitrella patens RAD51 genes is unique among eukaryotic organisms. Proc Natl Acad Sci USA 99: 2959–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Franklin AE, Golubovskaya IN, Bass HW, Cande WZ (2003) Improper chromosome synapsis is associated with elongated RAD51 structures in the maize desynaptic2 mutant. Chromosoma 112: 17–25. [DOI] [PubMed] [Google Scholar]
- 32. Pawlowski WP, Golubovskaya IN, Cande WZ (2003) Altered nuclear distribution of recombination protein RAD51 in maize mutants suggests the involvement of RAD51 in meiotic homology recognition. Plant Cell 15: 1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li W, Chen C, Markmann MU, Timofejeva L, Schmelzer E, et al. (2004) The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc Natl Acad Sci USA 101: 10596–10601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Markmann-Mulisch U, Wendeler E, Zobell O, Schween G, Steinbiss HH, et al. (2007) Differential requirements for RAD51 in Physcomitrella patens and Arabidopsis thaliana development and DNA damage repair. Plant Cell 19: 3080–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ayora S, Piruat JI, Luna R, Reiss B, Russo VEA, et al. (2002) Characterization of two highly similar Rad51 homologs of Physcomitrella patens . J Mol Biol 316: 35–49. [DOI] [PubMed] [Google Scholar]
- 36. Rajanikant C, Melzer M, Rao BJ, Sainis JK (2008) Homologous recombination properties of OsRad51, a recombinase from rice. Plant Mol Biol 68: 479–491. [DOI] [PubMed] [Google Scholar]
- 37. Sakane I, Kamataki C, Takizawa Y, Nakashima M, Toki S, et al. (2008) Filament formation and robust strand exchange activities of the rice DMC1A and DMC1B proteins. Nucleic Acids Res 36: 4266–4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kurumizaka H, Aihara H, Kagawa W, Shibata T, Yokoyama S (1999) Human Rad51 amino acid residues required for Rad52 binding. J Mol Biol 291: 537–548. [DOI] [PubMed] [Google Scholar]
- 39. Ishida T, Takizawa Y, Sakane I, Kurumizaka H (2008) The Lys313 residue of the human Rad51 protein negatively regulates the strand-exchange activity. Genes Cells 13: 91–103. [DOI] [PubMed] [Google Scholar]
- 40. Kagawa W, Kurumizaka H, Ikawa S, Yokoyama S, Shibata T (2001) Homologous pairing promoted by the human Rad52 protein. J Biol Chem 276: 35201–35208. [DOI] [PubMed] [Google Scholar]
- 41. Shibata T, DasGupta C, Cunningham RP, Radding CM (1979) Purified Escherichia coli recA protein catalyzes homologous pairing of superhelical DNA and single-stranded fragments. Proc Natl Acad Sci U S A 76: 1638–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McEntee K, Weinstock GM, Lehman IR (1979) Initiation of general recombination catalyzed in vitro by the recA protein of Escherichia coli . Proc Natl Acad Sci U S A 76: 2615–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bugreev DV, Mazin A (2004) Ca2+activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc Natl Acad Sci USA 101: 9988–9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Matsuo Y, Sakane I, Takizawa Y, Takahashi M, Kurumizaka H (2006) Roles of the human Rad51 L1 and L2 loops in DNA binding. FEBS J 273: 3148–3159. [DOI] [PubMed] [Google Scholar]
- 45. Endo M, Nakayama S, Umeda-Hara C, Ohtsuki N, Saika H, et al. (2012) CDKB2 is involved in mitosis and DNA damage response in rice. Plant J 69: 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang Z, Tang L, Li M, Chen L, Xu J, et al. (2010) Monitoring homologous recombination in rice (Oryza sativa L.). Mutat Res 691: 55–63. [DOI] [PubMed] [Google Scholar]
- 47. Wu Y, He Y, Moya IA, Qian X, Luo Y (2004) Crystal structure of archaeal recombinase RADA: a snapshot of its extended conformation. Mol Cell 15: 423–435. [DOI] [PubMed] [Google Scholar]
- 48. Chen Z, Yang H, Pavletich NP (2008) Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 453: 489–494. [DOI] [PubMed] [Google Scholar]
- 49. Aihara H, Ito Y, Kurumizaka H, Yokoyama S, Shibata T (1999) The N-terminal domain of the human Rad51 protein binds DNA: structure and a DNA binding surface as revealed by NMR. J Mol Biol 290: 495–504. [DOI] [PubMed] [Google Scholar]
- 50. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]