Fig 4.
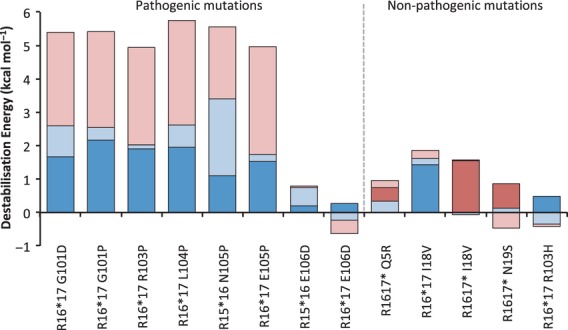
Pathogenic mutations are far more destabilizing than non-pathogenic mutations in the natural tandem repeat context. Thirteen mutations were created in both single-and multidomain contexts. Where the multidomain protein is R1516, ‘domain 1’ is R15; where the protein is R1617, ‘domain 1’ indicates R16. The ‘expected destabilization’ (domain 1, dark blue; domain 2, red) is the effect of the mutation on the single-domain protein. The ‘extra destabilization’ (domain 1, light blue; domain 2, pink) is the extra destabilization observed in the multidomain protein. Apart from E106D, all the pathogenic mutations result in loss of inter-domain stabilizing interactions. This results in an overall destabilization of > 5 kcal·mol−1. The non-pathogenic mutations all maintain the inter-domain interactions, and the loss in stability is essentially that ‘expected’ from the single-domain protein results, i.e. below 2 kcal·mol−1.
