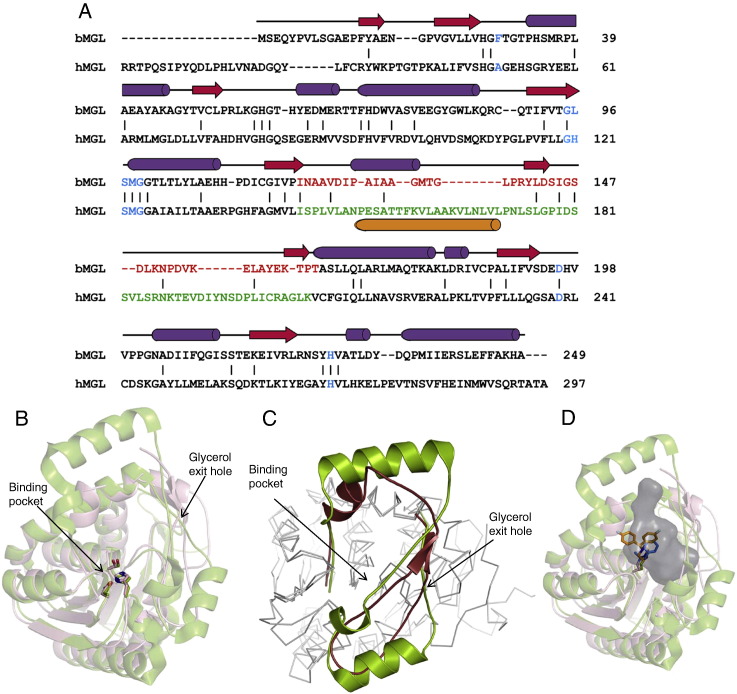
Fig. 7.
Structural comparison of bacterial and human MGL. A) Structure based sequence alignment of bMGL and hMGL (PDB ID: 3HJU) using Dali [58]. Secondary structure elements correspond to those observed in bMGL. β-strands and α-helices are depicted as red arrows and purple cylinders, respectively. Residues in blue correspond to the consensus G-X-S-X-G motif, the catalytic Asp and His, and the oxyanion hole forming residues. Residues within the cap regions of bMGL and hMGL are coloured in red and green, respectively. The proposed membrane binding helix α4 in the cap region of hMGL is shown as orange cylinder. B) Conservation of the catalytic triad evidenced by a structural superimposition. The structure of bMGL is shown as wheat brown cartoon and that of hMGL (PDB ID: 3JW8) as green cartoon. bMGL catalytic residues (Ser97-Asp196-His226) and hMGL (PDB ID: 3JW8) catalytic residues (Ser132-Asp249-His279) are shown as sticks in atomic colours. The substrate binding pocket and the proposed glycerol exit hole are indicated by arrows. C) Structural superposition of the cap region from bMGL (ruby red) and hMGL (green; PDB ID: 3JW8) shows a conserved architecture. D) bMGL (wheat brown) complexed to PMSF (blue) superimposed onto hMGL (green; PDB ID: 3JWE) complexed to SAR629 (orange). The cavity of bMGL as calculated by Casox is represented as a grey surface.