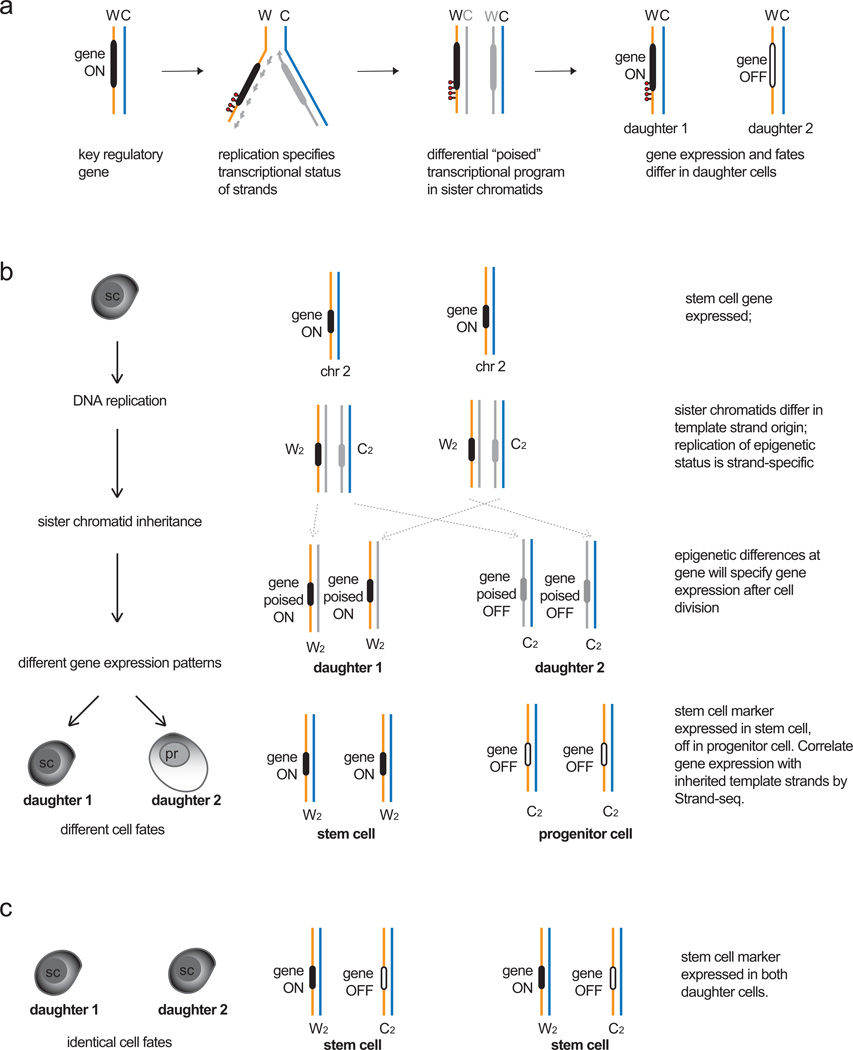
Figure 4.
The Silent Sister Hypothesis. a. Model for establishing different “poised” transcriptional states during DNA replication. For a given gene upon replication (black), strandspecific epigenetic marks (red) or epigenetic differences arising from leading strand (solid grey line) or lagging strand (dashed grey line) synthesis could lead to different poised epigenetic states between sister chromatids after replication is complete. Upon cell division, these differences could be translated as differential gene expression in the daughter cells. b. Differences in daughter cell identity could arise from inheritance of specific template strands and gene expression states in models of asymmetric cell division. If a stem cell expresses a key regulatory gene that specifies stem cell identity, then co-segregation of both Watson template strands encoding the gene and its expression status to the daughter stem cell (daughter 1) would maintain stem cell fate. Daughter 2 (progenitor, pr) no longer expresses the gene, therefore a differentiated fate program can be initiated. A symmetrical cell division could specify the same fate upon expression of the gene in both daughter cells. Strand-seq can identify patterns of template strand inheritance, which could then be correlated with gene expression patterns and cell fate programs in the daughter cells to test the Silent Sister Hypothesis.