Abstract
An Indian traditional fermented food, idli batter, was used as a source for isolation of lactic acid bacteria (LAB). A total of 15 LAB strains were isolated on the basis of their Gram nature and catalase activity. Of these, one lactobacilli strain and one lactococci strain which showed antimicrobial activity were identified using biochemical characterization, sugar utilization and molecular sequencing. The microbes, labeled as IB-1 (Lactobacillus plantarum) and IB-2 (Lactococcus lactis) were tested for their in vitro tolerance to bile salts, resistance to low pH values and acidifying activity. Both the strains showed good viability (IB1- 58.11%; IB2- 60.84%) when exposed to high bile salt concentration (2%) and acidic pH of ≤pH 3.0 (IB1- 88.13%; IB2- 89.85%). Lactic acid (IB1- 181.93 mM; IB2- 154.44 mM), biomass production (IB1- 0.65; IB2- 0.58 g/l) after 54 h as well as qualitative estimation of β-galactosidase and vitamin B12 production were also studied to check their suitability as an industrially important strain for production of important biomolecules.
Keywords: Idli batter, Probiotic, Lactic acid bacteria, Bile tolerance
Introduction
In recent decades, extensive research has been carried out on isolation and screening of microorganisms from traditional fermented foods due to their eco-friendly and genetically sturdy nature. Lactic acid bacteria (LAB) and yeasts play an important role in numerous natural food fermentations such as curd, cheese, pickles and various other traditional foods. Furthermore, they are closely associated with the human environment. LAB associated with fermented foods include species of the genera Enterococcus, Lactobacillus, Lactococcus, Leuconostoc, Pediococcus, Streptococcus, Tetragenococcus, Vagococcus and Weissella (Stiles and Holzapfel 1997). These organisms have also gained popularity as probiotics (Rivera-Espinoza and Gallardo-Navarro 2010). Probiotics are live microorganisms which, when taken in adequate amounts beneficially affect the host by improving the intestinal microbial balance (Fuller 1989). Among these microorganisms, LAB is regarded as a major group of probiotic bacteria (Collins et al. 1998).
A strain suitable as a probiotic should satisfy various criteria such as ability to resist acidic pH of stomach and bile salts produced at the opening of the intestinal tract, retain viability, and adhere to mucosal surfaces (Goldin and Gorbach 1992, Holzapfel et al. 1998). In addition, these strains should be non pathogenic and have a “generally regarded as safe” (GRAS) status. They should be able to grow easily and have a moderately good survival period. Their health benefits should also be well documented (Stanton et al. 2003). Generally probiotic cultures are used to reinstate the body’s naturally occurring gut flora after a course of antibiotics. Probiotics are used in controlling blood pressure and cholesterol levels, preventing gastrointestinal infection by pathogenic bacteria, treatment of diarrhoea and urogenital diseases (Reid 1999). Studies have also shown that probiotics strengthen the immune system to fight against allergies, stress, excessive alcohol intake and other diseases (Nichols 2007, Sanders 2003). Probiotics are additionally being scrutinized as live delivery medium to carry vaccines, antimicrobials or enzymes to targeted locations on the GI tract or mucosal surfaces. Isolated LAB’s are also used for production of lactic acid and polylactic acid (PLA). Fermentation is preferred for production of lactic acid due to favourable economics of production. PLA is a biodegradable material that has been used for orthopaedic implants, surgical sutures, disposable products and drug delivery systems (Adnan and Tan 2007).
Probiotic products have shown a rapid growth in the global food market due to increased consumer awareness on health and their demand for healthy foods. The cultures belonging to the genera Bifidobacterium, Lactobacillus, Lactococcuus and Saccharomyces, are used in many products ranging from fermented dairy products and infant food to pharmaceutical preparations.
A large number of LAB, isolated from various fermented food systems in different parts of the world have been studied for their probiotic potential and ability to produce industrially important substances. In India, idli batter fermentation has been the focus of research studies since a long time. A large number of LAB have been identified to be a part of the microflora responsible for fermentation of idli batter and they include Leuconostoc mesenteroides, Lactobacillus coryneformis, L.delbrueckii, L. fermentum, L.lactis, Streptococcus faecalis, and Pediococcus cerevisiae (Yajurvedi 1980). Areas such as preparation methods, nutritive value and microbiology of idli have been extensively studied and well documented (Desikachar et al. 1960; Steinkraus et al. 1967; Venkatasubbaiah et al. 1984; Kanchana et al. 2008; Riat and Sadana 2009; Sridevi et al. 2010). However, the probiotic potential of these microbes and their ability to produce industrially important substances has been seldom explored. Thus, in the current study isolates of LAB from idli batter (traditional Indian fermented food) were checked for their probiotic potential. Other health benefits and technological suitability of the isolates has also been studied.
Materials and methods
Materials
Black gram (Phaseolus mungo L.) and parboiled rice used for making idli batter were procured from local market. Bile salts (Bile acids, min 70%) was obtained from SD Fine Chemicals, Mumbai, India. Vitamin B12 standard was procured from Centron Research Laboratories, Mumbai, X-gal (5-bromo-4-chloro-3- indolyl-β-D-galactopyranoside) and IPTG (isopropyl- thio-β-D-galactopyranoside) was purchased from Bangalore Genei, India. ONPG was procured from Merck, Mumbai. E. coli NRRL 3008 was obtained from NRRL, Agricultural Research Service Culture Collection, Peoria, USA. Bacillus cereus MTCC 1272 and Listeria monocytogenes MTCC 1143 were purchased from MTCC, Chandigarh.E. coli NCIM 2068 for Vitamin B12 assay was procured from NCIM, Pune. MRS and nutrient agar medium was purchased from HiMedia Labs Pvt. Ltd, India. All other chemicals used were of analytical grade.
Strain isolation and identification
Idli batter preparation and isolation
The procedure for preparation of idli batter was as followed by Iyer and Ananthanarayan (2008). For isolation of LAB from idli batter, 10 g of the batter was added to 90 ml saline (0.9% w/v NaCl) and blended thoroughly on a orbital shaker for 30 min. Appropriate serial dilutions of the blended mixture was plated onto MRS agar (HiMedia Labs Pvt. Ltd., India) and incubated at 37°C for 48 h. This procedure was repeated at regular time intervals during the course of fermentation. Well isolated translucent/opaque colonies, 2–3 mm in diameter having entire margins were picked up and suspended in MRS broth and incubated at 37°C for 48 h. The process was repeated until pure cultures were obtained. These isolated LAB’s were maintained on MRS agar slants, by subculturing them periodically and stored in 40% glycerol at −20°C.
Identification of the idli batter isolates
The isolates obtained were checked for their Gram nature, morphology, catalase and antimicrobial activity. Two isolates exhibiting maximum antimicrobial activity (labeled as IB-1 and IB-2) were further characterized by physiological and biochemical tests according to Bergey’s Manual of Systematic Bacteriology (Mundt 1986). These included the study of fermentation type, agar plug test, triple sugar utilization test and citrate utilization test. Sugar utilization ability of the isolates was also studied. Finally, IB-1 and IB-2 were sequenced for 16S rRNA (National Centre for Cell Science, Pune) to confirm the results obtained from biochemical characterization. The sequences of the selected isolates were submitted to GenBank (NCBI).
Determination of antimicrobial activity
Antimicrobial activity was assayed by an adaptation of the critical dilution assay method (Mayr-Harting et al. 1972). The 48 h culture grown in MRS medium was centrifuged for 20 min at 4°C, 8,000 rpm and the supernatant thus obtained was adjusted to pH 5.5 (maximum antimicrobial activity was observed at pH 5.5) and used to determine antimicrobial activity. B. cereus MTCC 1272, E. coli NRRL 3008 and L. monocytogenes MTCC 1143 were used as indicator organisms. Nutrient agar 2% (10 ml) was overlaid with nutrient agar 1% (5 ml) inoculated with overnight grown culture suspensions of the indicator organisms. The plates were allowed to solidify and wells of 6 mm diameter were punched into them with a sterile cork borer. Cell free extract (100 μl) was poured in each of the wells and the plates were placed in the refrigerator at 4°C for 20 min to enhance diffusion of sample. The plates were then incubated at 37°C for 24 h and examined for zone of inhibition. The antimicrobial activity was determined by measuring the zone of clearance around the wells (well diameter + zone of inhibition). The activities of the cell free fractions were classified as no inhibition zone (−), inhibition zone of 6.5–7.5 mm (+), inhibition zone of 7.5–9.0 mm (++), and inhibition zone >9.0 mm (+++). Each assay was performed in triplicates.
Bile tolerance
Isolates IB-1 and IB-2 were grown in MRS broth containing 2% (w/v) of bile salts mixture at 37°C for 24 and 48 h. The growth was checked using the pour plate technique (Seeley and VanDemark 1971) wherein 1 ml of culture of appropriate dilutions was overlaid with MRS agar. The plates were incubated at 37°C for 48 h and the cell count was compared with that of the control MRS agar plates (containing cultures grown in MRS medium without bile salts mixture). Bacterial growth was expressed as colony forming units per milliliter (CFU/ml) and the survival percentage (% ± sd) of strains to bile salts was calculated as given below. (Mourad and Nour-Eddine 2006)
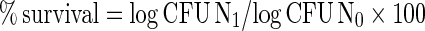 |
Where,
- N1
is viable count after exposure to bile salts
- N0
is viable count without exposure to bile salts
Tolerance to acidic pH values
Isolates IB-1 and IB-2 were grown in MRS broth at 37°C for 48 h. The cultures were centrifuged at 8,000 rpm for 10 min at 4°C. The pellets were washed twice in sterile phosphate-buffered saline (PBS), pH 7 and resuspended (1:100) in PBS to achieve a cell density of 1 × 1012 cells/ml. This was employed for setting up the experimental control and studying survival of isolates at low pH (pH 1, 2 and 3 prepared in PBS). The suspensions were incubated at 37°C and samples were removed after every 1 h up to 4 h. Counts of surviving cells were determined by plating on MRS agar using the procedure followed in bile tolerance assay. Bacterial growth was expressed in CFU/ml and the survival percentage (% ± sd) of strains to different pH values was calculated.
Lactic acid and biomass production
A time-course experiment to study the growth and lactic acid production profiles of IB-1 and IB-2 was carried out. The isolates were grown in 30 ml of basal MRS medium, pH 6.5 (in 100 ml flasks). Flasks were removed every 6 h up to 54 h. For biomass determination, 2 ml of the cells were centrifuged and washed twice with distilled water and kept for drying at 60°C until constant weight was obtained. The rest of the media was centrifuged and the broth obtained was used for measuring lactic acid content and pH. The lactic acid content was measured using a modified spectrophotometric method by Pryce (1969) and expressed in mM. The pH of the fermented broth was measured directly by using a digital Equip-Tronics ® pH meter (Mumbai, India).
Screening for vitamin B12 and β- galactosidase activity of IB-1 and IB-2
Vitamin B12 production was checked by plate method using E. coli NCIM 2068, a vitamin B12 auxotroph. The E. coli was grown for 18 h in Harrison medium containing (in g/l) glucose, 30; K2HPO4, 21; KH2PO4, 9; sodium citrate, 15; (NH4)2SO4, 3; asparganine; 0.6, MgSO4; 0.3 (Harrison et al. 1951). The flasks containing molten Harrison medium was inoculated at around 45°C with the 18 h old culture (O.D. 0.04 at 540 nm; 2% v/v) and immediately poured into sterile petri plates. The medium was allowed to solidify and 4 wells were punched in each of the plates using a sterile cork borer. Fermented broth (100 μl) was poured into two of the wells and a standard, filter sterilized Vitamin B12 solution (2–10 μg/ml) was poured into the other two wells. The plates were incubated at 37°C for 24 h after which the zone of exhibition was measured around the wells and compared with standard vitamin B12 solution (Atta et al. 2008)
For screening of β–galactosidase activity, the isolates were grown on MRS agar plates containing 0.01% X-gal (5-bromo-4-chloro-3- indolyl-β-D-galactopyranoside) and 0.1 mM IPTG (isopropyl- thioβ-D-galactopyranoside) as an inducer and the plates were incubated at 37°C for 48 h. Appearance of blue coloured colonies was indicative of β–galactosidase production (Karasová et al. 2002). For quantification of β–Galactosidase, the isolates were grown in modified MRS medium (glucose was replaced with lactose) at 37 ºC, 48 h. The bacterial cells were harvested by centrifugation at 8,000 rpm, 4°C for 15 min and washed twice in distilled water. The cells thus obtained were suspended in 0.2 M, pH 7 phosphate buffer and lysed by ultra sonication (Branson 450 Sonifier, Danbury, Connecticut., USA) set at 80% duty cycle, output 3 for 5 min to get a crude enzyme solution. The β–galactosidase activity of the abovementioned solution was determined using the o-NitroPhenylGalacto-Pyranoside (ONPG) method (Hestrin et al. 1976).
All the experiments were performed in triplicates (n = 3) and the difference (standard deviation) in the readings was less than or equal to ±5%.
Results and discussion
Strain isolation and identification
A total of 15 LABs were isolated at different time intervals during the course of fermentation of idli batter. Four bacterial isolates were obtained from black gram after soaking it for 4 h at 37°C. Six bacterial isolates were obtained from idli batter after 4 h of fermentation, and five bacterial isolates were obtained from batter after 16 h of fermentation. All the isolates were Gram positive in nature and catalase negative. Some of the cultures were bacilli (short rods), the others were cocci and one was a coccobacilli. Based on the Gram nature, morphology and catalase test, the cultures were observed to belong to the LAB family. One of the important features of a probiotic culture is its ability to kill pathogens which infect the gastrointestinal system. The isolates were also checked for their antimicrobial activity against B. cereus MTCC 1272, L. monocytogenes MTCC 1143 and E. coli NRRL 3008 which are commonly found food borne pathogens that infect the GI tract. The results showed that two of the fifteen isolates could inhibit the indicator organisms, however, at different inhibition levels. All these results have been compiled in Table 1. The isolate IB-2 showed antibacterial activity against all the three indicator organisms, whereas IB-1 showed activity against B. cereus MTCC 1272 and E. coli NRRL 3008. Several researchers have observed many of the LAB’s to be capable of producing antimicrobial substances that are active against pathogenic bacteria (Topisirovic et al. 2006). The differences in inhibition potential among the selected isolates could be due to different intrinsic factors induced by food origins (Klayraung et al. 2008).
Table 1.
General characteristics and antimicrobial activity of the isolated strains
| Isolate no. | Cell morphology | Antimicrobial activitya | ||
|---|---|---|---|---|
| E. coli | B. cereus | L.monocytogenes | ||
| 1 | Bacilli | + | + | − |
| 2 | Bacilli | + | + | − |
| 3 | Bacilli | + | + | − |
| 4 | Bacilli | + | + | − |
| 5 | Bacilli | + | ++ | − |
| 6 | Cocci | + | + | + |
| 7 | Bacilli | + | + | − |
| 8 | Cocci | ++ | + | − |
| 9 | Bacilli | + | + | − |
| 10 | Cocci | + | + | − |
| 11 (IB-1) | Bacilli | +++ | +++ | − |
| 12 (IB-2) | Cocci | +++ | +++ | ++ |
| 13 | Coccobacilli | + | + | − |
| 14 | Cocci | − | − | − |
| 15 | Bacilli | + | + | − |
aAntimicrobial activity: (−) no inhibition zone, (+) 6.5–7.5 mm inhibition zone, (++) 7.5-9 mm inhibition zone, (+++) >9 mm inhibition zone. 2 selected strains were labelled as IB-1 & IB-2. B. cereus MTCC 1272, E. coli NRRL 3008 and L. monocytogens MTCC 1143. (n = 3)
Biochemical characterization and sugar utilization studies of IB-1 and IB-2 was carried out (Tables 2 and 3). IB-1 was found to be heterofermentative while IB-2 was homofermentative. Both the isolates seemed to follow mixed type fermentation and could not utilize citrate as carbon source. The sugar utilization pattern showed that the isolates could belong to L. plantarum (bacilli) and L. lactis (cocci) (Mundt 1986). The two isolates were subjected to 16S rRNA sequencing and the cultures were found to have 99% sequence similarity to that of L. plantarum (IB-1) and L. lactis (IB-2). The sequences were uploaded in NCBI and have been given accession number GU797249 and GU797248 for IB-1 and IB- 2, respectively.
Table 2.
Biochemical characterization of the isolates IB-1 and IB-2
| Tests | Medium used | Results | |
|---|---|---|---|
| IB-1 | IB-2 | ||
| Fermentation type | Glucose phosphate broth | MR positive (Mixed acid fermentation) | |
| Agar plug test | Homo –Hetero fermentative medium | Gas formation (Heterofermentative) | No gas formation (homofermentative) |
| Triple sugar utilization | TSI agar slants | Yellow coloured slant and butt (Utilizes all sugars without H2S & gas. No utilization of N source after exhaustion of C source) | |
| Citrate utilization | Simmon’s citrate utilization agar tests | No change in colour (does not use citrate as carbon source) | |
(n = 3)
Table 3.
Carbon source utilization of the isolates L .plantarum (IB-1) and L .lactis (IB-2)
| Strain | Carbon source | Identified species | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mal | Xyl | Cel | Ara | Tre | Sal | Sor | Suc | Raf | Rha | Mant | Mann | Meli | Gal | ||
| IB-1 | + | V | V | V | + | + | + | + | + | + | + | + | + | + | Lactobacillus plantarum |
| IB-2 | + | − | V | + | + | + | − | + | − | − | − | + | − | V | Lactococcus lactis |
Mal maltose; Xyl xylose; Cel cellibiose; Ara arabinose; Tre trehalose; Sal salicilin; Sor sorbitol; Suc sucrose; Raf raffinose; Rha rhamnose; Mant mannitol; Mann mannose; Meli mellibiose; Gal galactose.
V Variable
(n = 3)
In vitro evaluation as a probiotic
Bile tolerance of IB-1 and IB-2
One of the important criteria to be fulfilled for a LAB to be used as a probiotic is its ability to resist the effect of bile salts in the gastrointestinal tract (Lee and Salminen 1995). However, there are no reports on the exact concentration to which a selected strain should be tolerant. The physiological concentration of bile salts in the small intestine is anywhere between 0.2 and 2.0% (Gunn 2000). Therefore the isolates were treated with 2% bile as it is the highest concentration obtained in animal and human intestine during digestion process (Gotcheva et al. 2002). Both the isolates showed good resistance (IB-1: 58.11% and IB-2: 60.84%) to 2% bile salt even after exposure for 48 h. Resistance to bile is related to bile salt hydrolase (BSH), an enzyme which helps in hydrolysing conjugated bile, thus reducing its toxic effect (Du Toit et al. 1998). This differs significantly among the LAB species and their strains. Similar results were also reported by Mourad and Nour-Eddine (2006) who found one of their isolated strains L. plantarum OL 16 to show 65% survival rate on exposure to 2% bile salt.
Acid tolerance of IB-1 and IB-2
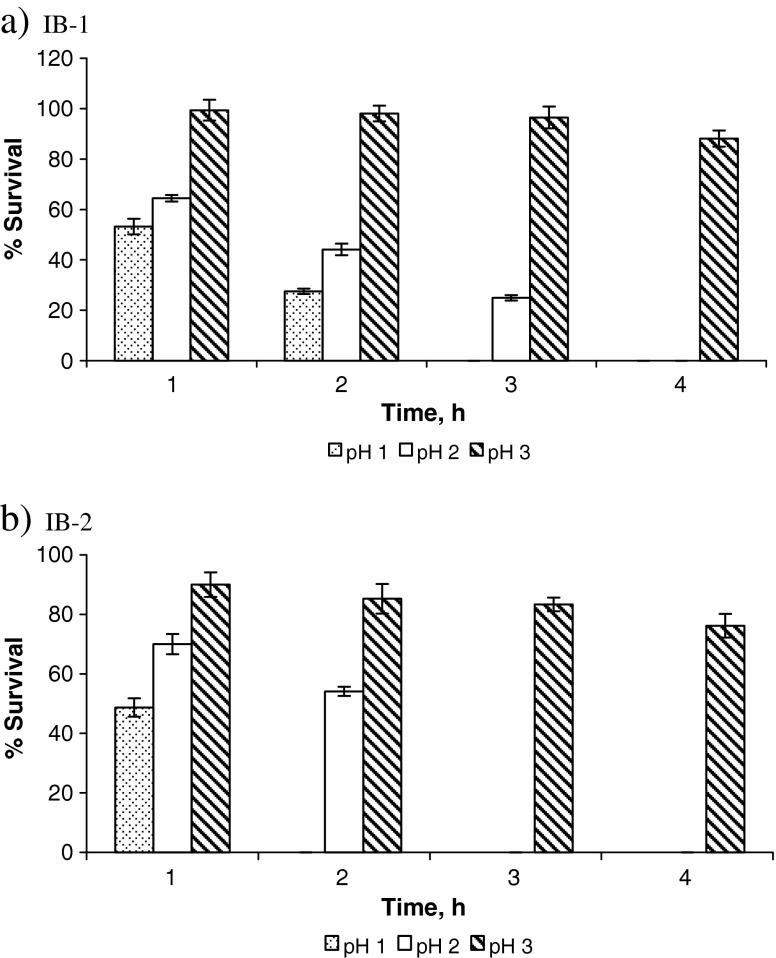
A probiotic strain should survive transit through the stomach where pH can be as low as 1.5 to 2. Hence, tolerance to extremely acidic conditions is another important feature of a probiotic strain (Dunne et al. 2001, Guo et al. 2009). Figure 1 summarizes the results of acid tolerance (survival percentage) of IB-1 and IB-2 at pH 1.0, 2.0 and 3.0 studied for a period of 1, 2, 3 and 4 h. Throughout the study, it was seen that IB-1 (L. plantarum) showed better survival as compared to IB-2 (L. lactis). It was observed that at pH 3.0, IB-1 and IB-2 showed survival of 88.13% and 89.85%, respectively, even after 4 h of incubation. However, it was noted that the percentage of survivors of both the isolates decreased with a decrease in pH. The isolate IB-1 (L. plantarum) showed higher survival up to 3 h (64.41%, 44.11%, and 24.96% at 1, 2 and 3 h of exposure, respectively) as compared to IB-2 (L. lactis) which survived only up to 2 h (69.98% and 54.41% at 1 and 2 h of exposure, respectively) at pH 2.0. At pH 1.0, IB-1 showed 53.17% survival after 1 h and 27.51% survival after 2 h; IB-2 showed 48.69% survival after 1 h. Mourad and Nour-Eddine (2006) had also reported their L. plantarum strains isolated from olives to show better survival rates at acidic pH as compared to other strains. Many researchers have attributed the acid tolerant nature of LAB to induction of H+-ATPase activity (Matsumoto et al. 2004, Ventura et al. 2004). Therefore, the difference in the acid tolerance of the isolates IB-1 and IB-2 might be related to the difference in H+-ATPase activity of the two strains. Both the isolates survived at upto 4 h at pH 3.0.
Fig. 1.
Tolerance of a IB-1 (L. plantarum) and b IB-2 (L. lactis) to acidic pH (n = 3)
Evaluation of the potential of IB-1 and IB-2 as an industrial strain
Biomass and lactic acid production by IB-1 and IB-2
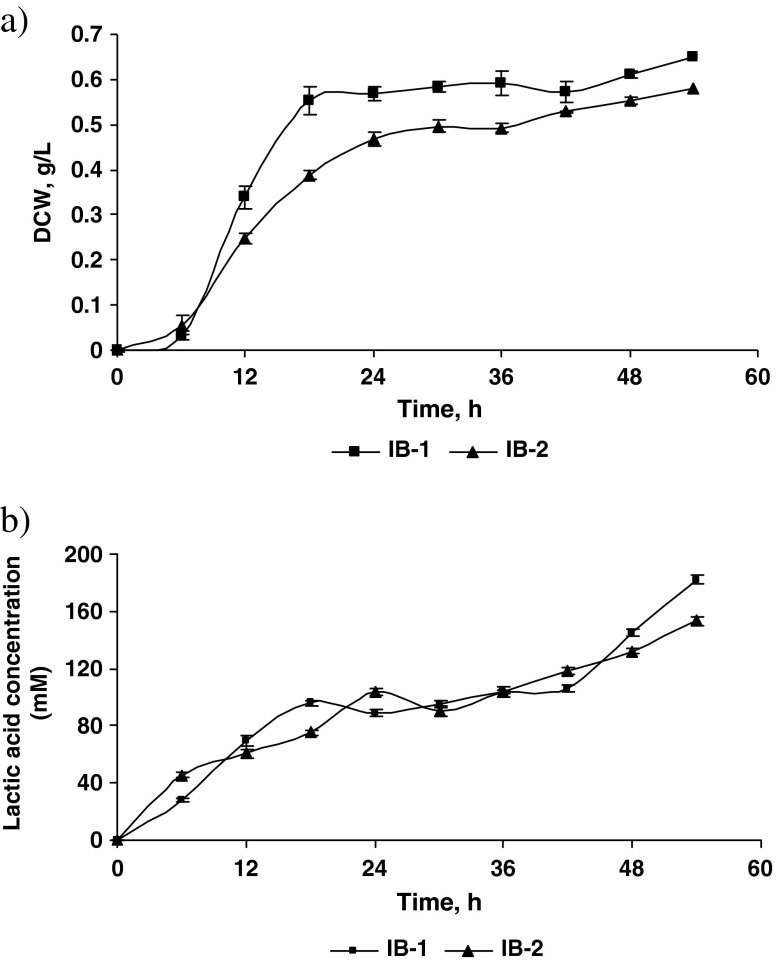
Figure 2 a & b depicts the biomass production in terms of dry cell weight (DCW) and lactic acid production (mM) profile of isolates IB-1 and IB-2. The time-course study was conducted to compare the growth and lactic acid producing capacity of lactobacilli and lactococci strain. The growth of both the isolates increased steadily upto 54 h at the end of which the DCW was 0.65 and 0.58 g/l for IB-1 and IB-2, respectively. It was observed that during the first 6 h, the lactococci strain IB-2 grew faster than the lactobacilli strain (IB-1). However, between 6 and 54 h the lactobacilli strain (IB-1) grew better than the lactococci strain (IB-2). A similar trend was observed in case of lactic acid production. The lactic acid content at the end of 54 h was 181.93 and 153.44 mM for IB-1 and IB-2 respectively. It was noticed that lactic acid production of both the isolates were parallel with that of biomass production. These results show that lactococci (IB-2) was less tolerant to lactic acid than lactobacilli (IB-1) and therefore there was lesser biomass produced in case of IB-2 strain. This confirmed the effect of lactic acid on biomass production. At industrial level, this problem could be solved by using a strategy wherein pH can be adjusted so as to help higher accumulation of lactic acid and biomass. A similar trend, wherein production of lactic acid limited growth of LAB was also observed by Adnan and Tan (2007). However in their study the amount of lactic acid produced was less and amount of biomass produced was higher, contrary to the results of our study.
Fig. 2.
a Biomass production (DCW/l) profile of the two isolates IB-1 and IB-2. b Lactic acid production (mM) profile of the two isolates IB-1 and IB-2 (n = 3)
Vitamin B12 and β-galactosidase production by IB-1 and IB-2
Vitamin B12 is an essential cofactor in amino acid, carbohydrate, fatty acid and nucleic acid metabolism, not produced by our body naturally but is required in good amounts for maintaining good health. It is predominantly of microbial origin, and is found in foods such as milk, meat and egg (Quesada-Chanto et al. 1994). In addition to dietary sources, intestinal microflora contributes to vitamin B12 in humans. A zone of exhibition of E. coli NCIM 2068 around the wells containing cell- free extracts of IB-1 and IB-2 broth indicated production of vitamin B12 by the isolates. A zone of growth was also observed surrounding the wells containing standard vitamin B12, as was expected. On comparing the zone of exhibitions of the standard and the sample it was seen that the broth contained approximately 8 μg/ml of Vitamin B12. Madhu et al. 2010 isolated an intracellular Vitamin B12 producing L. plantarum strain from Kanjika; however the selected isolates used in our study produced vitamin B12 extracellularly. No reports of an L. lactis strain producing Vitamin B12 have been reported so far. The reason behind the isolates producing Vitamin B12 maybe due to the presence of B12 biosynthetic gene cluster which encodes the enzymes required for the synthesis of this important vitamin (Santos et al. 2008). This could be probed further for industrial production of vitamin B12.
β-galactosidase, is an enzyme widely used in dairy industry. This enzyme is beneficial in preparation of lactose-free milk and biosynthesis of galactooligosaccharides that are important biomolecules from industrial and health point of view. In the current study it was observed that both the isolates IB-1 and IB-2 formed characteristic blue colored colonies due to hydrolysis of X-Gal. The ability of the isolates to hydrolyse X-gal (chromogenic substrate) in the medium qualitatively indicates the presence of β-galactosidase. Further these isolates when grown in liquid medium produced 0.5 U/ml of the enzyme. Researchers have reported presence of β-galactosidase activity in LAB (Aysun and Candan 2003). Evidences support the claim of lactose-intolerant individuals being able to tolerate fermented lactose rich dairy products better; these beneficial effects were attributed to microbial β-galactosidases (Vasiljevic and Shah 2008). These strains could be further explored for production of the enzyme.
Conclusion
L. plantarum (IB-1) and L. lactis (IB-2) strains isolated from fermented idli batter could tolerate high bile salt concentration and low pH. Based on these in vitro tests, there is high possibility that the isolates would be able to reach the intestinal tract in good numbers. Both the isolates were good lactic acid producers and also showed antibacterial activity against pathogenic microorganisms. The ability of the isolates to produce vitamin B12 and β -galctosidase was an added advantage as both of these are essential in improving digestion and metabolism. This could be considered a positive trait for microorganisms which are used as starter cultures and in manufacturing of probiotic and novel functional foods. However, the isolated strains need to be further investigated using in vivo experiments to establish their potential health benefits.
Acknowledgement
We would like to thank Prof. Yogesh Souche, National Centre for Cell Science, Pune University for helping in determining the 16S rRNA sequences of the isolated strains.
References
- Adnan AFM, Tan IKP. Isolation of lactic acid bacteria from Malaysian foods and assessment of the isolates for industrial potential. Bioresour Technol. 2007;98:1380–1385. doi: 10.1016/j.biortech.2006.05.034. [DOI] [PubMed] [Google Scholar]
- Atta HM, Arafa RA, Salem MS, El-Meleigy MA. Microbiological studies on the production of Vitamin B12 from two mixed cultures under solid state fermentation condition. J Appl Sc Res. 2008;4:1463–1477. [Google Scholar]
- Aysun C, Candan G. Properties of potential probiotic Lactobacillus plantarum strains. Food Microbiol. 2003;20:511–518. doi: 10.1016/S0740-0020(02)00174-0. [DOI] [Google Scholar]
- Collins JK, Thornton G, Sullivan GO. Selection of probiotic strains for human applications. Int Dairy J. 1998;8:487–490. doi: 10.1016/S0958-6946(98)00073-9. [DOI] [Google Scholar]
- Desikachar HSR, Radhakrishnamurthy R, Ramarao G, Kadkol SB, Srinivasan M, Subrahmanyan V. Studies on idli fermentation. I: Some accompanying changes in the batter. J Sci Ind Res. 1960;196:168–172. [Google Scholar]
- Du Toit M, Franz C, Schillinger U, Warles B, Holzappfel W. Characterization and selection of probiotic Lactobacilli for a preliminary minipig-feeding trail and their effect on serum cholesterol level, faeces moisture contents. Int J Food Microbiol. 1998;40:93–104. doi: 10.1016/S0168-1605(98)00024-5. [DOI] [PubMed] [Google Scholar]
- Dunne C, O’Mahony L, Murphy L, Thornton G, Morrissey D, O’Halloran S, Feeney M, Flynn S, Fitzgerald G, Daly C, Kiely B, O’Sullivan GC, Shanahan F, Collins JK. In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am J Clin Nutr. 2001;73:386S–392S. doi: 10.1093/ajcn/73.2.386s. [DOI] [PubMed] [Google Scholar]
- Fuller R. A review: Probiotics in man and animals. J Appl Bacteriol. 1989;66:365–378. doi: 10.1111/j.1365-2672.1989.tb05105.x. [DOI] [PubMed] [Google Scholar]
- Goldin B, Gorbach S. Probiotics for humans. In: Fuller R, editor. Probiotics—The scientific basis. London: Chapman and Hall; 1992. pp. 335–376. [Google Scholar]
- Gotcheva V, Hristozova E, Hristozova T, Guo M, Roshkova Z, Angelov A. Assessment of potential probiotic properties of lactic acid bacteria and yeast strains. Food Biotechnol. 2002;16:211–225. doi: 10.1081/FBT-120016668. [DOI] [Google Scholar]
- Gunn JS. Mechanisms of bacterial resistance and response to bile. Microbes Infect. 2000;2:907–913. doi: 10.1016/S1286-4579(00)00392-0. [DOI] [PubMed] [Google Scholar]
- Guo Z, Wang J, Yan L, Chen W, Liu X, Zhang H. In vitro comparison of probiotic properties of Lactobacillus casei Zhang, a potential new probiotic, with selected probiotic strains. LWT Food Sci Technol. 2009;42:1640–1646. doi: 10.1016/j.lwt.2009.05.025. [DOI] [Google Scholar]
- Harrison E, Lees KA, Wood F. The assay of vitamin B12. VI. Microbiological estimation with a mutant of Escherichia coli by the plate method. Analyst. 1951;76:696–705. doi: 10.1039/an9517600696. [DOI] [Google Scholar]
- Hestrin S, Feingold DS, Schram M. Lactose reduction of milk by fiber-entrapped beta-galactosidase. In: Mauro P, Franko M, editors. Methods in enzymology, vol. 1. New York: Academic; 1976. pp. 822–830. [DOI] [PubMed] [Google Scholar]
- Holzapfel W, Haberer P, Snel J, Schillinger U, Huis in’t Veld J. Overview of gut flora and probiotics. Int J Food Microbiol. 1998;41:85–101. doi: 10.1016/S0168-1605(98)00044-0. [DOI] [PubMed] [Google Scholar]
- Iyer BK, Ananthanarayan L. Effect of a-amylase addition on fermentation of idli—A popular south Indian cereal—Legume-based snack food. LWT Food Sci Technol. 2008;41:1053–1059. doi: 10.1016/j.lwt.2007.07.004. [DOI] [Google Scholar]
- Kanchana S, Raghavan GSV, Venkatesh S, Yuan G. Quality assessment of dehydrated Idli. J Food Sci Technol. 2008;45:177–179. [Google Scholar]
- Karasová P, Spiwok V, Malá Š, Králová B, Russell NJ. Beta-galactosidase activity in psychrotrophic microorganisms and their potential use in food industry. Czech J Food Sci. 2002;20:43–47. [Google Scholar]
- Klayraung S, Viernstein H, Sirithunyalug J, Okonogi S. Probiotic properties of Lactobacilli isolated from Thai traditional. Food Sci Pharm. 2008;76:485–503. doi: 10.3797/scipharm.0806-11. [DOI] [Google Scholar]
- Lee YK, Salminen S. The coming age of probiotics. Trends Food Sci Technol. 1995;6:241–245. doi: 10.1016/S0924-2244(00)89085-8. [DOI] [Google Scholar]
- Madhu AN, Giribhattanavar P, Narayan MS, Prapulla SG. Probiotic lactic acid bacterium from kanjika as a potential source of vitamin B12: evidence from LC-MS, immunological and microbiological techniques. Biotechnol Lett. 2010;32:503–506. doi: 10.1007/s10529-009-0176-1. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Ohishi H, Benno Y. H+-ATPase activity in Bifidobacterium with special reference to acid tolerance. Int J Food Microbiol. 2004;93:109–113. doi: 10.1016/j.ijfoodmicro.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Mayr-Harting A, Hedges AJ, Berkeley RCW. Methods for studying bacteriocins. In: Norris JR, Ribbons DW, editors. Methods in microbiology, 7A. New York: Academic; 1972. pp. 315–422. [Google Scholar]
- Mourad K, Nour-Eddine K. In vitro preselection criteria for probiotic Lactobacillus plantarum strains of fermented olives origin. Int J Probiotics Prebiotics. 2006;1:27–32. [Google Scholar]
- Mundt JO. Lactobacillus. In: Sneath PHA, Mair NS, Sharpe ME, Holt JG, editors. Bergey’s manual of systematic bacteriology. Baltimore: Williams & Wilkins; 1986. pp. 577–592. [Google Scholar]
- Nichols AW. Probiotics and athletic performance: a systematic review. CurrSports Med Rep (Current Medicine Group LLC). 2007;6:269–273. [PubMed] [Google Scholar]
- Pryce JD. A modification of the Barker-Summerson method for the determination of lactic acid. Analyst. 1969;94:1151–1152. doi: 10.1039/an9699401151. [DOI] [PubMed] [Google Scholar]
- Quesada-Chanto A, Afschar AS, Wagner F. Microbial production of propionic acid and Vitamin B12 using molasses or sugar. Appl Microbiol Biotechnol. 1994;41:378–383. doi: 10.1007/BF00939023. [DOI] [PubMed] [Google Scholar]
- Reid G. The scientific basis for probiotic strains of Lactobacillus. Appl Environ Microbiol. 1999;65:3763–3766. doi: 10.1128/aem.65.9.3763-3766.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riat P, Sadana B. Effect of fermentation on amino acid composition of cereal and pulse based foods. J Food Sci Technol. 2009;46:247–250. [Google Scholar]
- Rivera-Espinoza Y, Gallardo-Navarro Y. Non-dairy probiotic products. Food Microbiol. 2010;27:1–11. doi: 10.1016/j.fm.2008.06.008. [DOI] [PubMed] [Google Scholar]
- Sanders ME. Probiotics: considerations for human health. Nutr Rev. 2003;61:91–99. doi: 10.1301/nr.2003.marr.91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, Vera JL, van der Heijden R, Valdez G, de Vos WM, Sesma F, Hugenholtz J. The complete coenzyme B12 biosynthesis gene cluster of Lactobacillus reuteri CRL1098. Microbiol. 2008;154:81–93. doi: 10.1099/mic.0.2007/011569-0. [DOI] [PubMed] [Google Scholar]
- Seeley HW Jr, VanDemark PJ (1971) Microbes in action: A laboratory manual of microbiology pour plates 2nd edn, p. 22.
- Sridevi J, Halami PM, Vijayendra SVN. Selection of starter cultures for idli batter fermentation and their effect on quality of idlis. J Food Sci Technol. 2010;47:557–563. doi: 10.1007/s13197-010-0101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton C, Desmond C, Coakley M, Collins JK, Fitzgerald G, Ross RP. Challenges facing development of probiotic-containing functional foods. In: Farnworth ER, editor. Handbook of fermented functional foods. Florida: CRC; 2003. pp. 27–58. [Google Scholar]
- Steinkraus KH, Van Veck AG, Theireau DB. Studies on idli-an Indian fermented black gram rice food. Food Technol. 1967;21(6):110–111. [Google Scholar]
- Stiles ME, Holzapfel WH. Lactic acid bacteria of foods and their current taxonomy. Int J Food Microbiol. 1997;36:1–29. doi: 10.1016/S0168-1605(96)01233-0. [DOI] [PubMed] [Google Scholar]
- Topisirovic L, Kojic M, Fira D, Golic N, Strahinic I, Lozo J. Potential of lactic acid bacteria isolated from specific natural niches in food production and preservation. Int J Food Microbiol. 2006;112:230–235. doi: 10.1016/j.ijfoodmicro.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Vasiljevic T, Shah NP. Probiotics—from Metchnikoff to bioactives. Int Dairy J. 2008;18:714–728. doi: 10.1016/j.idairyj.2008.03.004. [DOI] [Google Scholar]
- Venkatasubbaiah P, Dwarakanath CT, Sreenivasa Murthy V. Microbiological and physicochemical changes in idli batter during fermentation. J Food Sci Technol. 1984;22:59–63. [Google Scholar]
- Ventura MD, van Sinderen G, Fitzgerald F, Zink R. Insights into the taxonomy, genetics and physiology of bifidobacteria. Antoine van Leeuwenhoek. 2004;86:205–233. doi: 10.1023/B:ANTO.0000047930.11029.ec. [DOI] [PubMed] [Google Scholar]
- Yajurvedi RP. Microbiology of idli fermentation. Indian Food Packer. 1980;34:33–36. [Google Scholar]