Figure 3.
Characteristics of SNPs Identified from RNA-Seq Data of GM12878 Cells
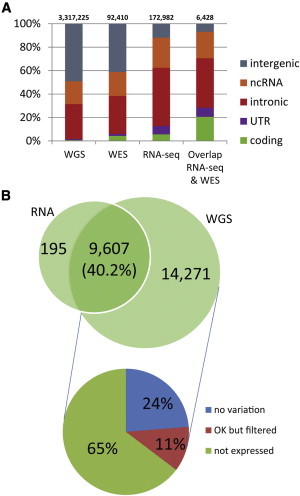
(A) The composition of genomic regions for variants in WGS, WES, and RNA-seq suggests a high enrichment of RNA-seq variants in functionally important regions. Sites present in RNA-seq and WES occurred substantially more often in coding exons.
(B) Overlap in coding variants detected from RNA-seq and WGS. Of all coding variants, 40.2% were found by RNA-seq. The majority of the remaining sites were not detected as a result of the lack of expression. “No variation” indicates that the position was homozygous in RNA, “OK but filtered” indicates that the position was heterozygous but was removed by one of our filtering steps, and “not expressed” indicates that the position was not covered by RNA-seq reads.
