Figure 1.
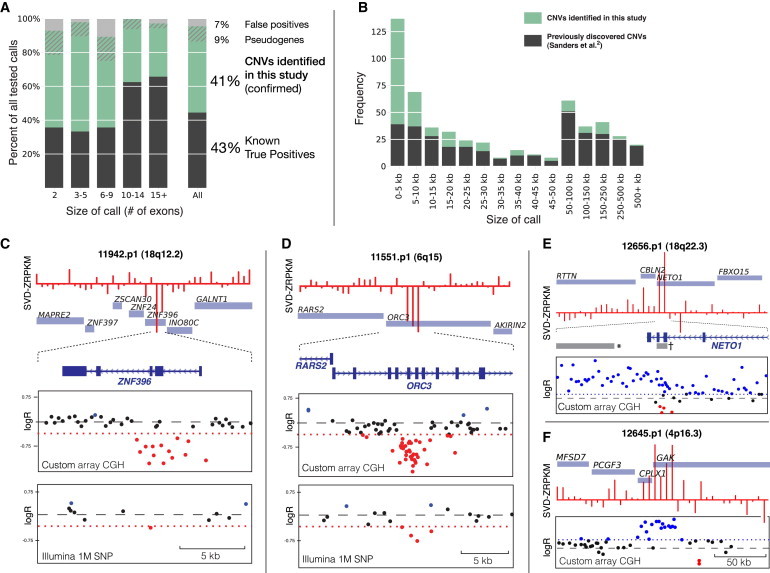
Discovery and Validation of CNVs with the Use of Exomes
(A) Fraction of CNVs previously identified via Illumina 1M SNP microarray (gray, “known true positives”), the fraction of CNVs identified and confirmed by targeted array CGH in this study (green, “CNVs identified in this study”), confirmed processed pseudogenes (hatched green), and the overall FPR for unconfirmed CNVs (gray).
(B) The majority (73% [152/207]) of all calls (green) identified in this study with the use of exomes were smaller than 20 kb.
(C–D and F) Three examples of CNVs in this study. Top: CoNIFER output and normalized coverage at each exon. Middle: targeted array CGH at CNV locus; the threshold for deletion or duplication (dotted red line) was determined by ROC-curve analysis of known CNVs (Supplemental Data). Bottom: Illumina 1M SNP microarray data for locus shows poor probe coverage (C and D only).
(E) Exome-based CNV discovery affords high exon-level specificity, as indicated by duplication of NETO1 exons (†, CoNIFER call). Previous work (Sanders et al.2) discovered this CNV (∗), but the (incorrect) breakpoints did not extend into NETO1.
