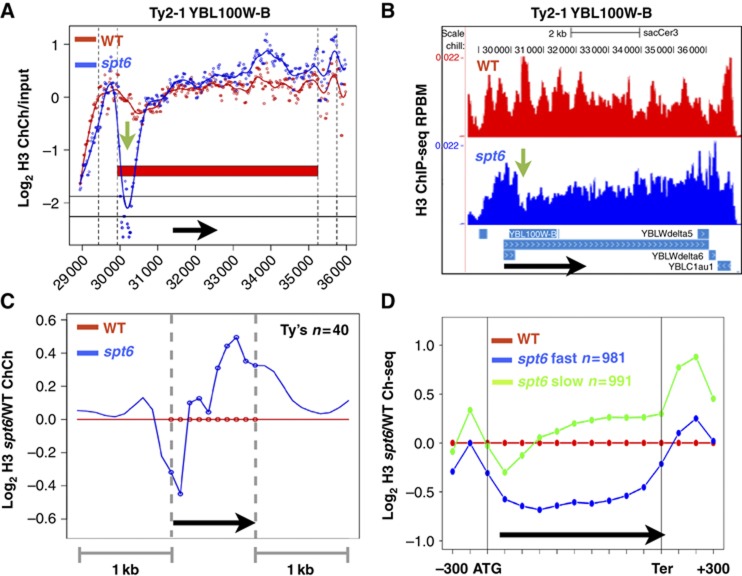
Figure 4.
(A, B) Localized loss of H3 over a region of 1–2 nucleosomes (green arrows) at the 5′ end of Ty2-1 in independent ChIP-Chip and ChIP-seq experiments. Note that ChIP-Chip signals at repetitive sequences will be affected by cross-hybridization. Reads per base pair per million mapped reads (RPBM). (C) Average distributions of histone H3 ChIP-Chip signals for the spt6-td degron relative to WT at 40 full-length Ty elements (Supplementary Table S1). (D) Average distributions of histone H3 ChIP-seq signals in the spt6-td degron and WT strains (37°C+Dox). The 1000 genes with the fastest and slowest exchange rates averaged over the ORF and 5′ and 3′ flanking regions (Rufiange et al, 2007) are analysed (Supplementary Table S1). Note greater eviction on fast exchanging genes when Spt6 is inactivated.