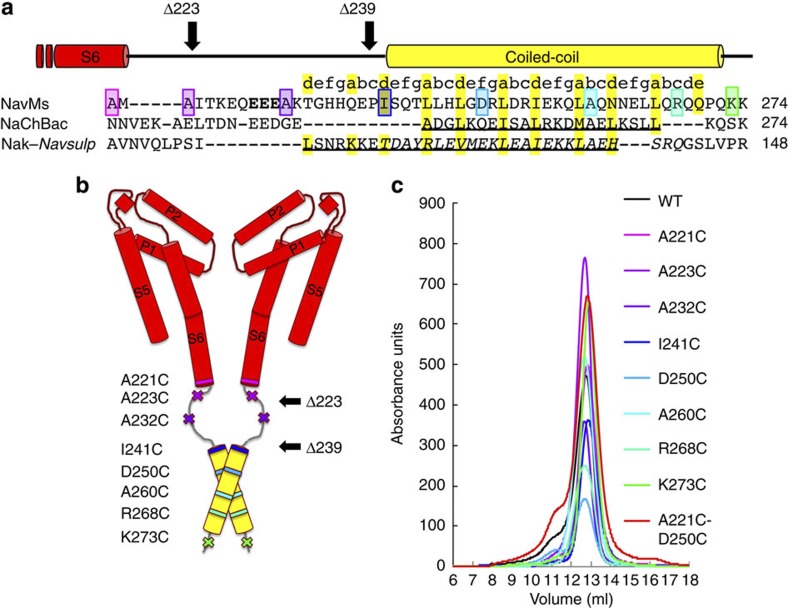
Figure 1. Labelled proteins.
(a) Alignment of the CTDs of bacterial sodium channels starting at the end of transmembrane helix S6 (red bar above alignment). GenBank nucleotide accession codes: NavMs, YP_864725; NaChBac, NP_242367; chimeric NaK-NavSulP, with NaK, AB617622 and (italicised) NavSulP, NAS-14.1, AALZ01000002. The coiled-coil region predicted for NavMs is indicated by a yellow bar above the sequence alignment. The residues in NaChBac, which form the helical region5 and the region of the NaK-NavSulP chimera, which forms a coiled-coil14 are underlined. Residues that were mutated to cysteines and spin labelled are in coloured boxes. The same colouring scheme for each mutant is used in all figures: transmembrane residue A221 is pink, linker region residues A223 and A232 are lilac and purple, respectively; residues I241, D250, A260 and R268 in the predicted coiled-coil region are shades of blue and the final residue K273 is green. The EEE residues changed in the QQQ mutant used for electrophysiology are in bold. (b) Model of the open pore channel, adapted from McCusker et al.9 showing the locations of the spin-labelled amino acids. The outward displacement of the end of transmembrane helix S6 in this open form is due to a kink at T209 in the middle of S6. Only two of the four monomers are shown for clarity. The black arrows indicate the positions of the truncations (Δ223 and Δ239) used in the electrophysiology experiments. (c) Size exclusion chromatography of wild-type and mutant proteins used in the EPR experiments, showing all run as well-behaved tetramers.