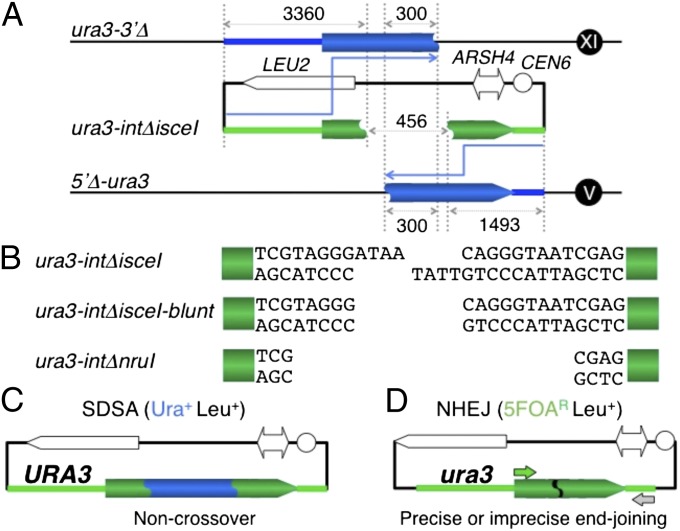
Fig. 1.
SDSA/NHEJ assay. (A) For this assay, two different donor-alleles, 5′Δ-ura3 [with a deletion from the promoter (the 221-nt upstream of the initiation codon) to the first base of the 39th codon, residing at the ura3 locus on Chr. V] and ura3-3′Δ [the ura3 ectopic allele bearing only the region from the promoter (the 221-nt upstream of the initiation codon) to the first base of the 139th codon of the URA3 gene, integrated into the AUR1 locus on Chr. XI], were constructed (13). The plasmids with the recipient alleles, ura3-intΔnruI and ura3-intΔisceI (the internal 458-bp deletion of the URA3 gene, sealed with an NruI site and an I-SceI site, respectively, residing within the plasmid with the LEU2 marker), were introduced into the double-template strains with the ura3-3′Δ allele and 5′Δ-ura3 allele, which share 300-bp internal homology. The homologous regions between ura3-intΔ and ura3-3′Δ or 5′Δ-ura3 are 3,360 bp and 1,493 bp in length, respectively. (B) ura3-intΔisceI, ura3-intΔisceI-blunt and ura3-intΔnruI generate 4-nucleotide 3′ tails by I-SceI cleavage for pSDSA/NHEJ (with the I-SceI site) plasmid DNA, blunt ends by T4 DNA polymerase treatment following the I-SceI cleavage, and blunt ends by NruI cleavage for pSDSA/NHEJ (with the NruI site) plasmid DNA, respectively. (C) SDSA products as Ura+ Leu+ transformants bearing the URA3 plasmids (13). The 458-bp gap is repaired via SDSA. (D) NHEJ products as Ura− (5FOAR) Leu+ transformants bearing the ura3 plasmids (13). The retention or deletion of the I-SceI sequence was detected by PCR with the primers (arrows) (13), I-SceI nuclease treatment, and sequence determination as precise or imprecise end-joining, respectively (Table 1).