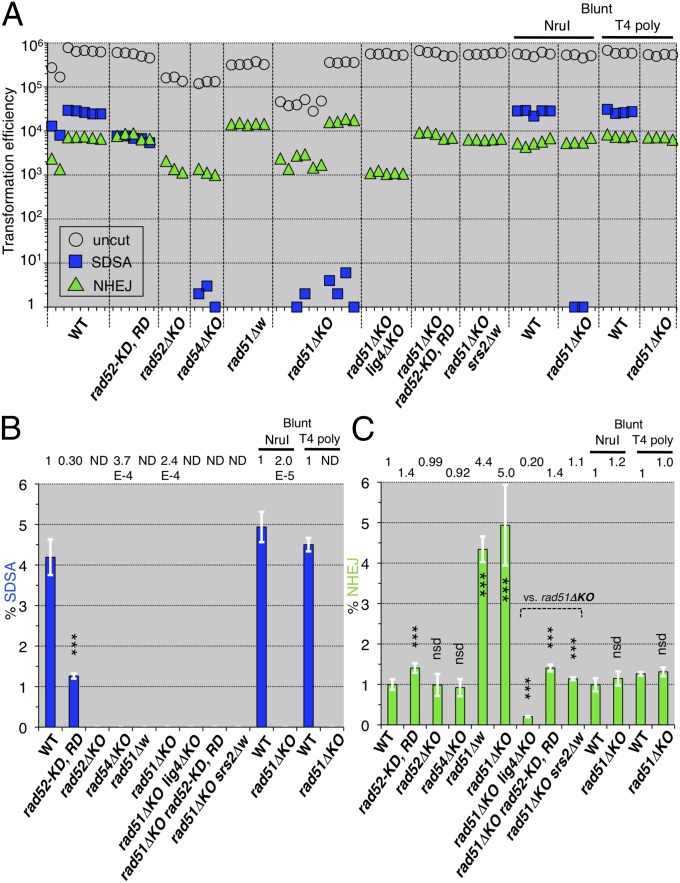
Fig. 2.
SDSA and NHEJ of homologous recombination-deficient mutants. (A) Leu+ transformation efficiencies with the uncut plasmid bearing ura3-intΔisceI are plotted as the transformation competencies of the cell suspensions (open circles). Ura+ Leu+ and 5-FOAR Leu+ transformation efficiencies with the I-SceI–cut plasmid bearing ura3-intΔisceI are plotted as numbers of SDSA progeny (blue squares) and as numbers of NHEJ progeny (green triangles), respectively (Fig. 1). (B) The normalized frequencies (%) of SDSA events were calculated from the transformation efficiencies (Materials and Methods) and plotted (blue bars). “ND” indicates that no SDSA progeny were detected. (C) The normalized frequencies (%) of the NHEJ events were calculated from the transformation efficiencies (Materials and Methods) and plotted (green bars). rad51ΔKO lacks most of the coding region, and rad51Δw lacks the entire coding region, from the initiation codon to the last sense codon (Tables S1 and S2). rad52-KD, RD shows rad52-K117D, R148D. “NruI” and “T4 poly” indicate the use of the blunt-ended ura3-intΔnruI and ura3-intδΔisceI-blunt DNAs, respectively. The statistical analysis (Table S3): nsd, no significant difference; *P < 0.05; **P < 0.01; ***P < 0.001; two-tailed Student’s t test (vs. wild-type except for vs. rad51ΔKO); error bars = SD.