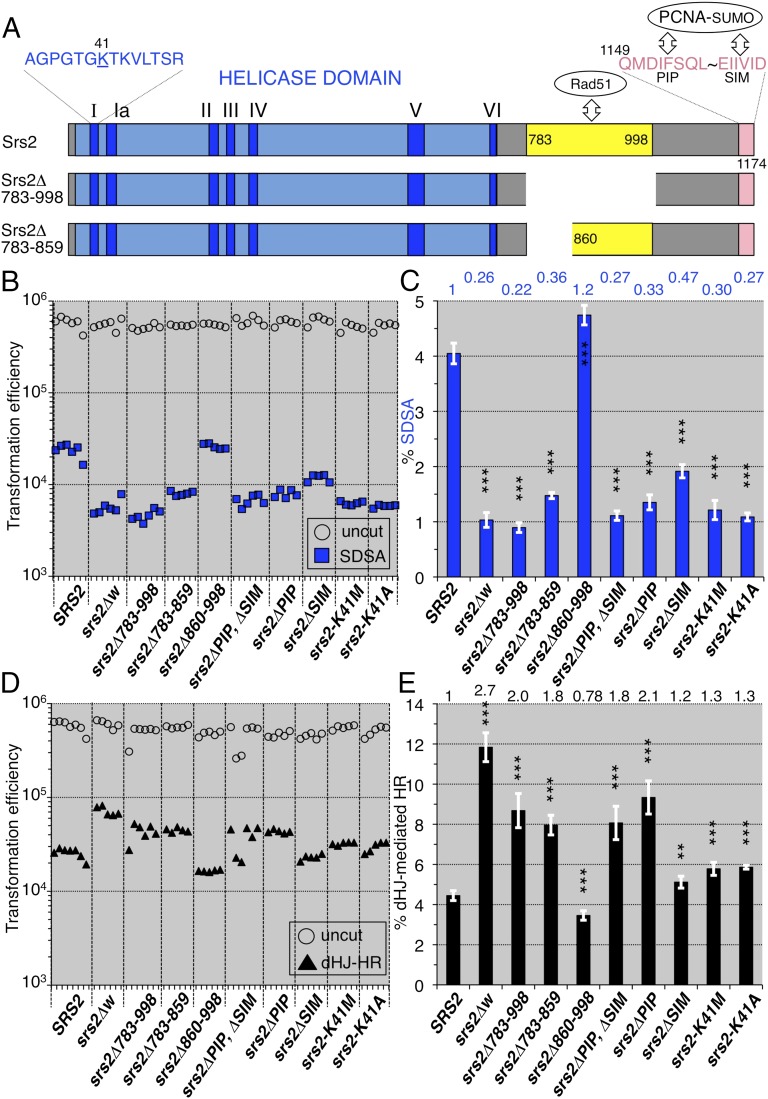
Fig. 3.
SDSA and double Holliday junction-mediated homologous recombination (dHJ-mediated HR) of various srs2 mutants. (A) The Srs2 helicase domain (blue box), including seven consensus motifs (dark blue), and the locations of the Rad51-binding domain (783-998), the N-terminal half of the Rad51-binding domain (783-859), PIP motif (1149-1156), and SIM motif (1169-1174). The 41st residue (lysine) is substituted with methionine and alanine in the srs2-K41M and srs2-K41A mutants, respectively. (B) Leu+ and Ura+ Leu+ (SDSA progeny) transformation efficiencies with uncut and I-SceI–cut plasmids possessing ura3-intΔisceI, respectively. (C) The normalized frequencies (%) of the SDSA events were calculated from the transformation efficiencies (Materials and Methods) and plotted (blue bars). (D) Targeted integration completed by double Holliday junction-mediated homologous recombination (Fig. S2). The plot shows the AurR transformation efficiencies with the uncut pRS315-AurR plasmid as the transformation competencies of the cell suspensions (open circles), and the AurR transformation efficiencies with the StuI-cut pAUR101 plasmid as the numbers of double Holliday junction-mediated homologous recombination progeny (closed triangles). (E) The normalized frequencies (%) of the double Holliday junction-mediated homologous recombination were calculated from the transformation efficiencies (Materials and Methods) and plotted (black bars). Statistical analysis (Tables S4 and S5): nsd, no significant difference; *P < 0.05; **P < 0.01; ***P < 0.001; two-tailed Student’s t test (vs. wild-type); error bars = SD.