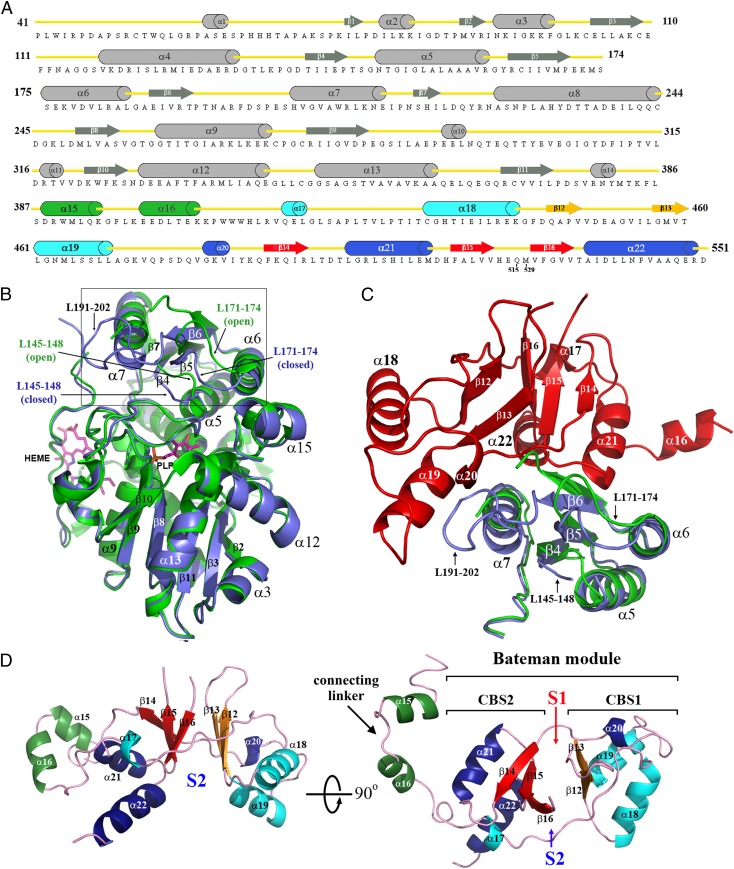
Fig. 2.
Topology and structure of hCBSOPTΔ516–525. (A) Cartoon presentation of the secondary structures within the C-hCBSOPTΔ516–525 amino acid sequence. (B) Structural superimposition of the truncated 45-kDa hCBS protein (green; PDB ID code 1JBQ) with the catalytic core of the full-length hCBS protein (blue). The loops L145–148, L171–174, and L191–202 are located at the entrance of the PLP cavity and regulate the access of substrates into the catalytic site. As depicted, the loop L191–202 is disordered in the truncated 45-kDa hCBS protein and thus is not visible. The open conformation adopted by the loops L145–148 and L171–174 in the absence of the regulatory domain explain why the truncated enzyme is activated. (C) Interface between the catalytic core (blue) and the Bateman module (red) in the basal form of the full-length enzyme. The truncated 45-kDa hCBS protein (green) is superimposed for comparison. As depicted, the Bateman module pushes the entrance loops toward the PLP cavity, thus hindering the access of substrates. (D) The 3D structure of the Bateman module, which contains two tandem-repeated CBS motifs (CBS1, residues 412–471, and CBS2, residues 477–551). Secondary structure elements have been colored as in A. S1 and S2 designate the proposed AdoMet-binding sites.