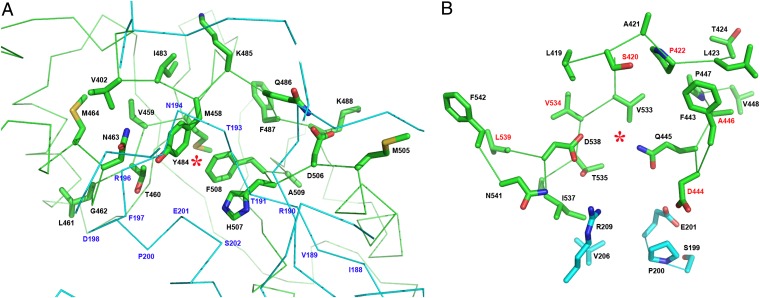
Fig. 5.
Putative AdoMet-binding sites in hCBS. (A) Site S1. The entrance of S1 is sterically hindered by the presence of structural elements from the catalytic core (cyan) of a complementary monomer in the dimer. Additionally, bulky hydrophobic residues (Y484, H507, and F508) occupy the cavity and probably impede the binding of AdoMet at this site in the basal form. Residue N463, instead of the conserved aspartate that would stabilize the nucleotide ribose ring, could play a similar role. (B) Site S2. In contrast, the cavity of S2 is solvent exposed and is not blocked by bulky residues. The site S2 shows features similar to the AdoMet-binding protein MJ0100 (27): a hydrophobic cage to host the adenine ring of AdoMet, conserved aspartate (D538) and threonine (T535) residues to stabilize the ribose ring, and a hydrophobic residue (I537) preceding D538 to accommodate the alkyl chain of AdoMet. Noteworthy are the residues linked with pathogenic mutations (identified in red).