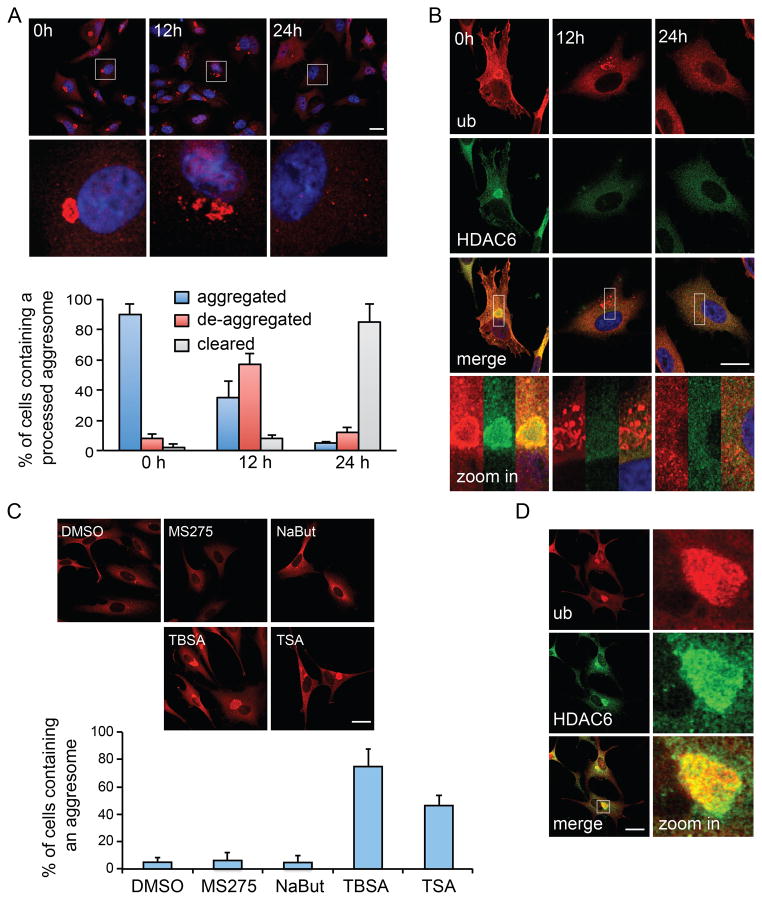
Figure 1. HDAC6 is required for aggresome de-aggregation and clearance.
A549 cells were treated with MG132 (5 μM) for 24h to induce aggresome formation, followed by MG132-free medium for the indicated times. Aggresomes were identified by staining with an antibody against ubiquitin (ub, clone FK1). (A) Ubiquitin immuno-staining (red) indicates that the large aggresomes (0h) were de-aggregated (12h) before final clearance (24h). Bottom panels show zoomed areas (white squares). Nuclei were stained with DAPI (blue). The status of aggresomes at different time points after MG132 washout was quantified in the histogram. (B) Representative images of HDAC6 (green) in relationship to the aggresome (red) by double-immuno-staining during MG132 washout. Bottom panels show zoomed areas (white squares). Note that protein aggregates are no longer positive for HDAC6 after 12h washout (middle panels). (C) A549 cells were treated with an HDAC6-selective inhibitor, Tubastatin A (TBSA, 10 μM), pan-HDAC inhibitor, Trichostatin A (TSA, 1 μM), or class I HDAC inhibitors, MS275 (10 μM) or sodium butyrate (NaBut, 1mM) during MG132 washout. The presence of aggresome was analyzed and quantified as described in (A). (D) Representative images of HDAC6 and the aggresome (ub) in TBSA-treated cells. For (A) and (C), three independent experiments were quantified. Error bars show ± S.E.M. Scale bar = 25 μm. See also Figure S1.