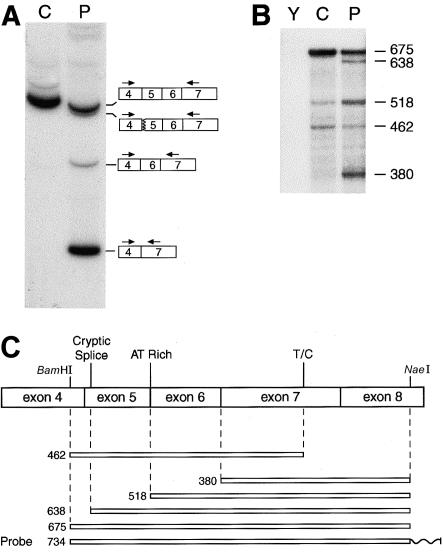
Figure 1.
Detection of abnormalities between exons 4 and 7 sequences in RNA from one of the patient's COL5A1 alleles. A, Amplification of COL5A1 cDNA from a control individual (C) and the patient (P) with a sense primer, end-labeled with γ[32P]-ATP (see the “Patient, Material, and Methods” section), in exon 4 and with an antisense primer in exon 7. A single major band is apparent in the sample from the control individual; the sample from the patient has the normal-sized band derived from the normal COL5A1 allele, one band just below that representing fragments produced by use of two separate cryptic acceptor sites in exon 5 (removing 12 or 15 nt from exon 5), and two lower bands that result from skipping of either exon 5 or both exons 5 and 6. B, Quantitative RNase-protection analysis. Conditions for labeling and annealing of the antisense probe to yeast (Y), control (C), and patient (P) RNA are described in the “Patient, Material, and Methods” section. Nuclease-protected fragments were separated on a denaturing 6% polyacrylamide gel. Sizes (in nt) of protected fragments are given. C, Schematic depicting the relationship of the riboprobe and protected fragments to COL5A1 exon sequences. BamHI and NaeI sites used in the making of the probe—as well as a cryptic splice site used in allele B, an AT-rich region that may result in partial cleavage by RNase due to “breathing” of the RNA duplex, and an SNP at nucleotide 1318 (T/C)—are shown. A wavy line represents non-COL5A1 vector sequences on the riboprobe.