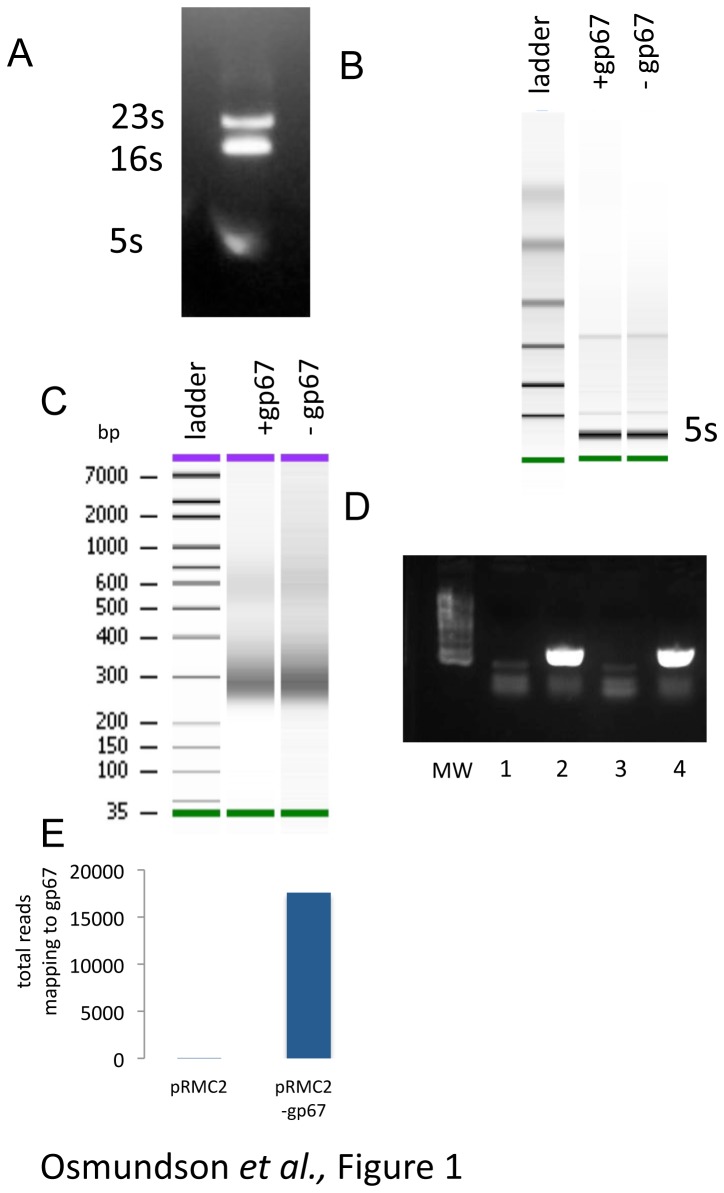
Figure 1. Schematic of RNA-seq in S. aureus.
a) Total RNA is purified from cells and verified for integrity on a 1% agarose gel. b) rRNA reduction is used to remove the large (16s and 23s) rRNA species from the sample. RNA was assessed by running the samples on a BioAnalyzer. c) After rRNA reduction, the standard Illumina random-prime technique was used to prepare a cDNA library for sequencing. DNA was assessed by running the samples on a BioAnalyzer. d) To verify the representation of mRNA in the cDNA library, and that the prepared samples differed predictably, we performed a PCR for cDNA corresponding to gp67. A band corresponding to gp67 cDNA is only present in cells containing pRMC2-gp67 (lane 4) and not control cells containing pRMC2 alone (lane 3). e) RNA-seq reads mapping to the gene for gp67. RNA-seq reads mapping to gp67 are only present in the RNA-seq data from cells containing pRMC2-gp67 and not control cells containing pRMC2 alone.