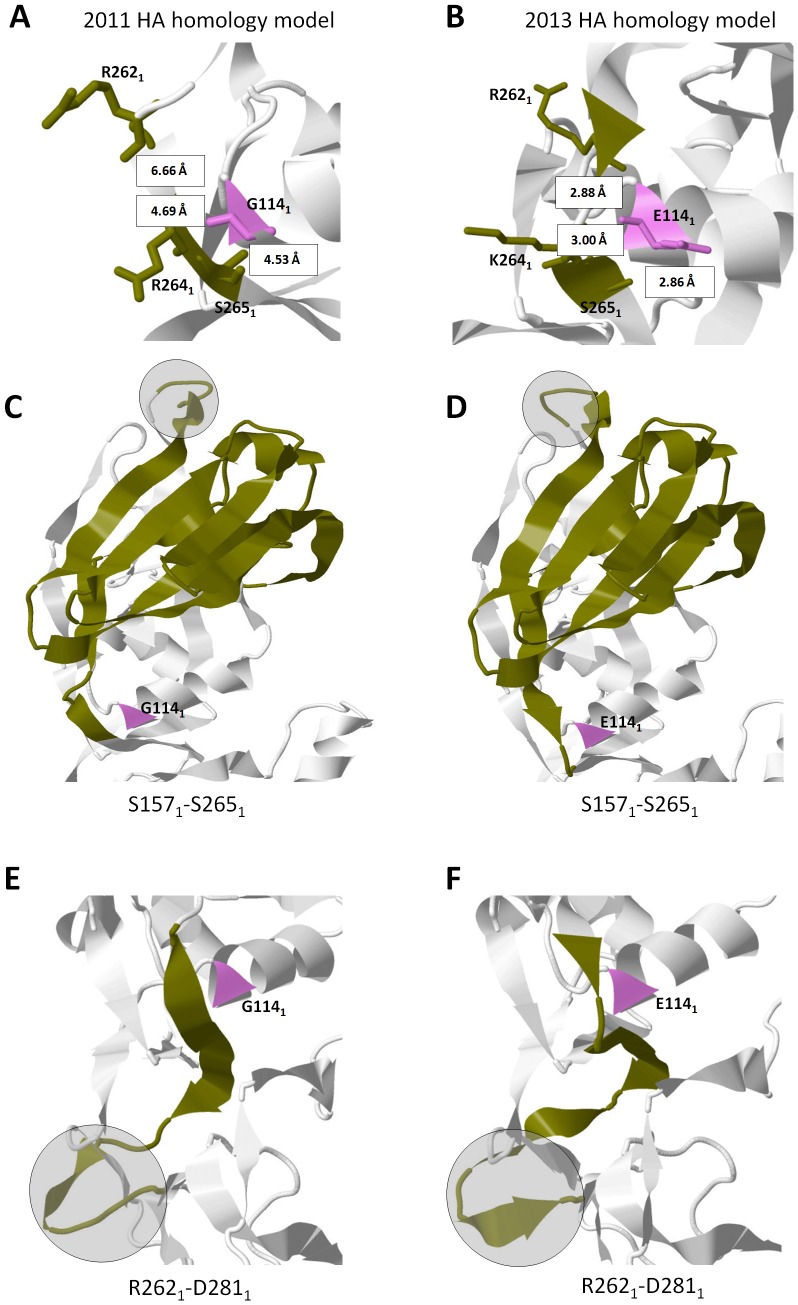
Figure 3. Pattern-1 structural differences are ascribable to salt bridge formation involving amino acid residue 1141.
Distance measurements of amino acid residues 2621, 2641, and 2651 (green) relative to residue 1141 (violet) found in the (A) 2011 and (B) 2013 HA homology models. All amino acid residues indicated are in a wireframe structure. All measurements are indicated in Å. Interrelationship of HA structural differences (shaded in gray) observed in the (C,E) 2011 and (D,F) 2013 HA homology models are highlighted in green. Amino acid residue 1141 is indicated in violet.