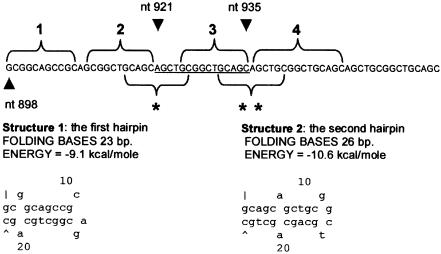
Figure 3.
Explanation of the mechanism of the novel triplication 921–935trip15 by a replication error. Predictions of DNA hairpins are based on MFOLD (Walter et al. 1994). When the DNA polymerase is stalling because of the GC richness of the region, the sequences 1 and 2 of the newly synthesized strand form a DNA hairpin because they are self-complementary (structure 1). This allows the sequence 3 to hybridize with the template of the region 2, since they have exactly the same sequence, and this leads to a duplication of the underlined sequence (nt 921–935). Once this extra sequence is present (4), there may be a rearrangement of the hairpin, which leads to the formation of another one involving the sequences (single and double asterisk). The formation of the second hairpin is favorable, because it is more stable (by an energy excess of 1.5 kcal/mole; see structure 2). Finally, the 3′ end of the sequence 4 hybridizes with the template of sequence 2, which results in the triplication after escaping repair. Further, notice that sequences 1–4 form a very stable, almost perfect, hairpin (−28 kcal/mole).