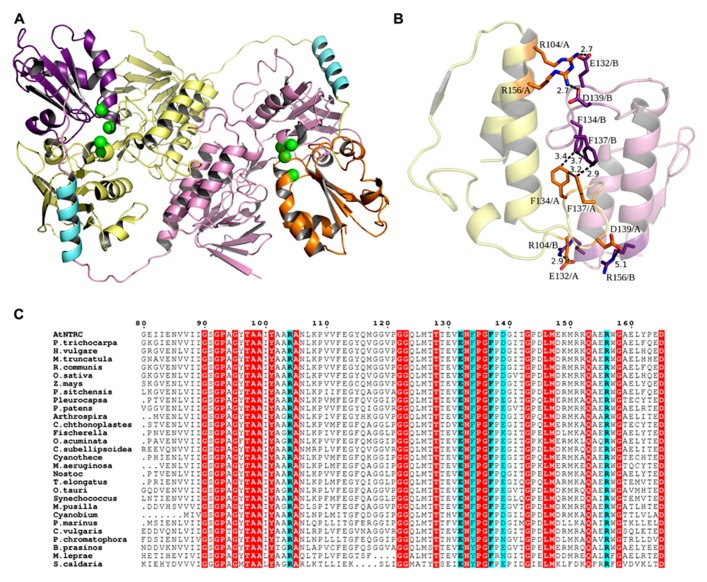
FIGURE 8.
(A) Dimer model of NTRC. In the dimer, the TRX domain (orange and purple) of one subunit binds to the NTR domain (yellow and pink) of the other subunit and, thus, tightens the inter-monomeric interactions. Within one subunit, a long linker region that starts from the C-terminus of the NTR domain and forms a short α-helix (cyan) connects to the N-terminus of the TRX domain, suggesting possible flexibility for the position of the TRX domain. Active site cysteines are shown as green spheres. (B) Dimer interactions. In addition to the arrangement of subunits and the linker-mediated interactions, five possible salt bridges are formed between the FAD binding domains. Four of these are shown as sticks in the picture (orange and purple), and three of these are at an optimal distance (~2.7 Å) from each other. Additionally, four phenylalanines in the middle of the interface interact with each other (pi-pi stacking) in a square. Altogether, these special interactions can account for the experimentally observed strong dimeric interactions. (C) NTRC like sequences. A part of the multiple sequence alignment of NTRC like sequences from Arabidopsis thaliana (UniProtKB O22229), Populus trichocarpa (UniProtKB B9H9S9), Hordeum vulgare (UniProtKB B0FXK2), Medicago truncatula (UniProtKB A6XJ27), Ricinus communis (UniProtKB B9SU44), Oryza sativa Japonica Group (UniProtKB Q70G58), Zea mays (GenBank DAA63960.1), Picea sitchensis (UniProtKB B8LPW6), Pleurocapsa sp. PCC 7327 (UniProtKB K9T492), Physcomitrella patens subsp. patens (UniProtKB A9U311), Arthrospira sp. PCC 8005 (UniProtKB H1WE73), Coleofasciculus chthonoplastes PCC 7420 (UniProtKB B4VQ87), Fischerella sp. JSC-11 (UniProtKB G6FX47), Oscillatoria acuminata PCC 6304 (UniProtKB K9TN60), Coccomyxa subellipsoidea C-169 (UniProtKB I0Z4C2), Cyanothece sp. PCC 7822 (UniProtKB E0U9J8), Microcystis aeruginosa PCC 7941 (UniProtKB I4GKJ6), Nostoc sp. PCC 7120 (UniProtKB Q8YYV6), Thermosynechococcus elongatus BP-1 (UniProtKB Q8DHM2), Ostreococcus tauri (UniProtKB Q00SZ2), Synechococcus sp. PCC 6312 (UniProtKB K9RZN2), Micromonas pusilla CCMP1545 (UniProtKB C1N9L2), Cyanobium sp. PCC 7001 (UniProtKB B5INL4), Prochlorococcus marinus subsp. marinus str. CCMP1375 (UniProtKB Q7VB53), Chlorella vulgaris (UniProtKB B9ZYY5), Paulinella chromatophora (UniProtKB B1X3G8), Bathycoccus prasinos (UniProtKB K8EJ46), Mycobacterium leprae TN (UniProtKB P46843) and Spirochaeta caldaria DSM 7334 (UniProtKB F8F2K7) show that the residues forming salt bridges of optimal length (cyan) are mostly conserved between species. Red boxes indicate other conserved residues.