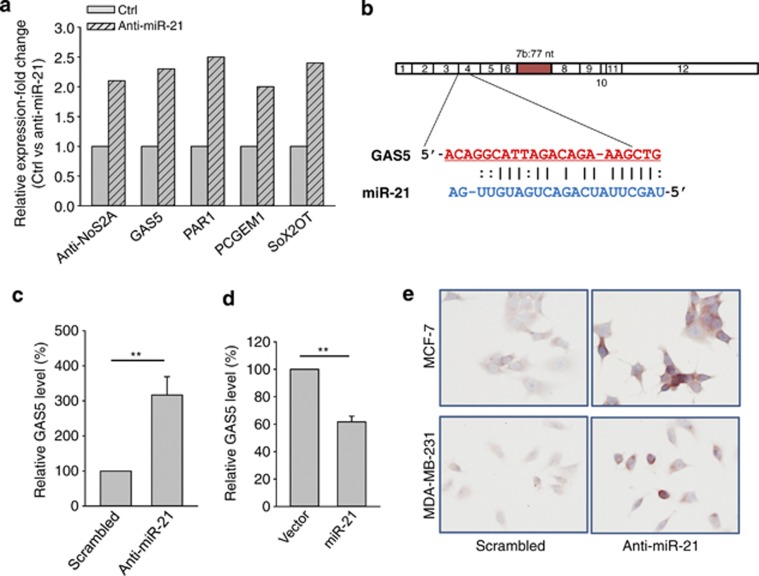
Figure 1.
Identification of GAS5 as an miR-21 target. (a) Identification of putative miR-21-regulated lncRNAs. MCF-7 cells were transfected with either scrambled oligo (Ctrl) or anti-miR-21. The cells were harvested 24 h after transfection and total RNA was isolated, followed by lncRNA profiling as described in the text. GAS5 was among five lncRNAs with over a two-fold increase by anit-miR-21. (b) Alignment of potential miR-21 base pairing with GAS5 as identified by RNA22 program (http://cbcsrv.watson.ibm.com/rna22.html). GAS5 (top) consists of 12 exons, where the putative binding site is in exon 4. The isoform b of GAS5 has 77 nucleotides at exon 7. (c) Detection of induction of GAS5 in the anti-miR-21 cells using newly designed primers (GAS5-RT-5.1 and GAS5-RT-3.1). Experimental procedure was same as in (a). (d) Ectopic expression of miR-21 suppresses GAS5. MCF-7 cells were transfected with vector alone or miR-21, and total RNA was isolated 24 h after transfection for RT-PCR. (e) Induction of GAS5 by anti-miR-21 as detected by in situ hybridization. MCF-7 or MDA-MB-231 cells were transfected with scrambled oligo or anti-miR-21. GAS5 signal was detected by in situ hybridization as detailed in the Materials and Methods section. Error bars represent S.E.M., n=3. **P<0.01