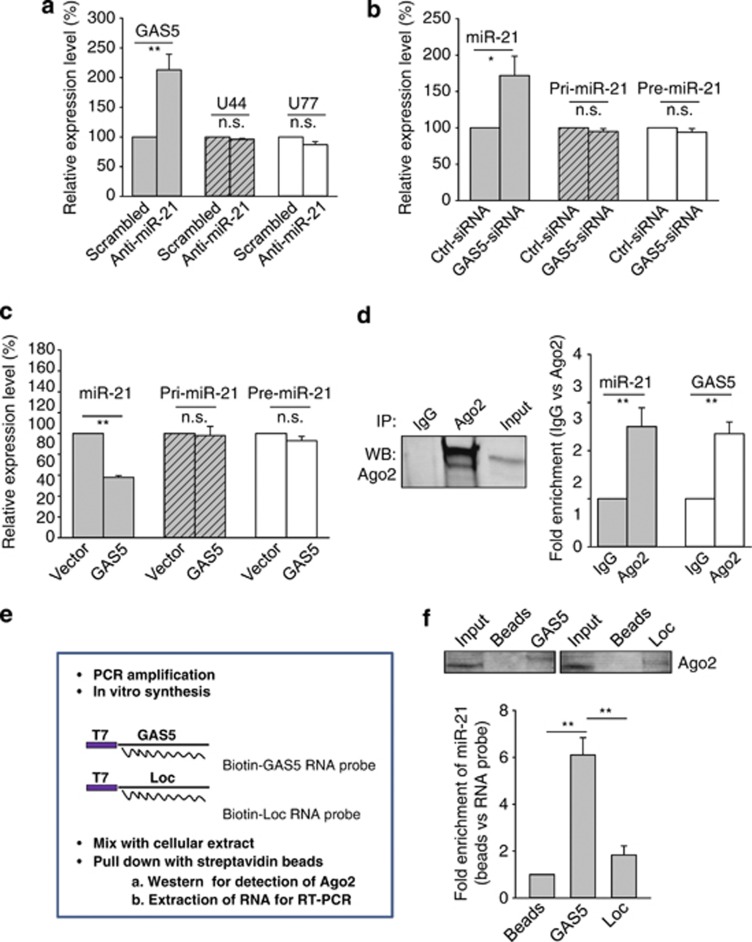
Figure 7.
Post-transcriptional regulation of GAS5 and miR-21. (a) Whereas anti-miR-21 upregulates GAS5, it has no effect on U44 and U77, which are imbedded in the GAS5 gene. MCF-7 cells were transfected with scrambled oligo or anti-miR-21 and the rest of the procedure was same as in Figure 1c. Primers for U44 and U77 were U44-RT-5.1 and U44-RT-3.1; U77-RT-5.1 and U77-RT-3.1, respectively. (b and c) Effect of ectopic expression of GAS5 or GAS5–siRNA on pri-miR-21, pre-miR-21 and mature miR-21. MCF-7 cells were transfected with vector alone or GAS5, or control siRNA or GAS5–siRNA. Total RNA was isolated from the cells 24 h later, followed by qRT-PCR. Primers for pri-miR-21 were Pri-miR-21-RT-5.1 and Pri-miR-21-RT-3.1; Pre-miR-21-RT-5.1 and Pre-miR-21-RT-3.1. (d) Association of GAS5 and miR-21 with AGO2. Cellular lysates from MCF-7 cells were used for RNA immunoprecipitation with AGO2 antibody. Detection of AGO2 using IP-western (left panel), and detection of GAS5 and miR-21 using qRT-PCR (right panel). (e and f) Identification of GAS5 and miR-21 in the same RISC complex by RNA precipitation. (e) A procedure for making biotin-labeled RNA probes and RNA precipitation. In vitro transcribed RNA probes were made using T7 RNA polymerase, followed by precipitation assays as described in the Materials and Methods section. (f) Detection of miR-21 in the GAS5 RNA-precipitated samples. Error bars represent S.E.M., n=3. *P<0.05; **P<0.01; n.s., not significant