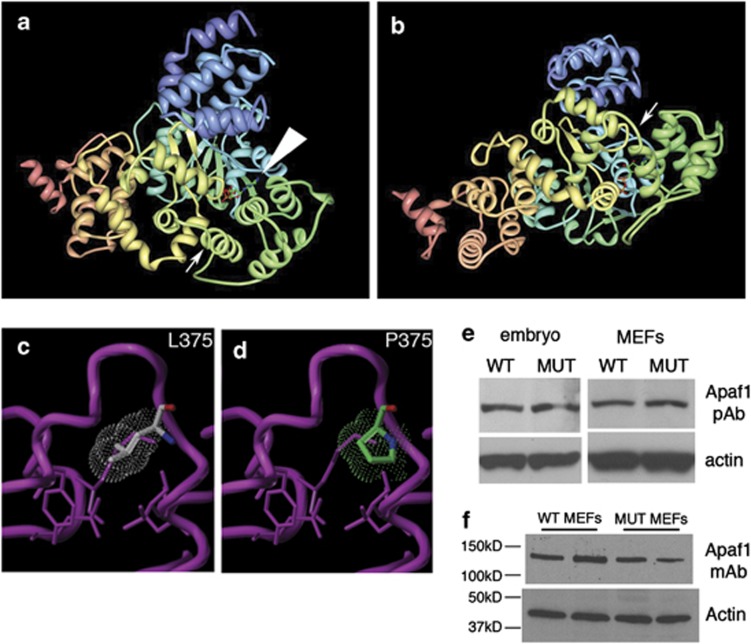
Figure 3.
The yautja mutation is predicted to disrupt the structure of Apaf1, but protein size and expression appear intact in mutants. (a) Image from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB; http://www.pdb.org) of PDB ID 1Z6T,17 with small arrow indicating the location of the yautja L375P mutation. Large arrowhead indicates the bound ATP molecular structure. (b) Same image as in a, but rotated 90° away from viewer, to show the yautja mutation location in the loop between α-helices in the same orientation as depicted in the close-up models (c and d). Close-up of the winged-helix domain in wild-type (WT; L375) (c) and mutant (MUT; P375) (d) versions of Apaf1 showing steric hindrance (dot cloud) on the protein backbone (purple pipe/ribbon) generated by proline substitution (Sybyl models provided by Kim Gernert at Emory University School of Medicine Biomolecular Computing Resource). (e and f) Apaf1 protein is stably expressed in Apaf1yautja mutant embryos and MEFs. Western blot of whole-cell lysates from WT and Apaf1yautja MUT embryos or embryonic fibroblasts (MEFs) using either polyclonal (e) or monoclonal (f) antibodies against Apaf1